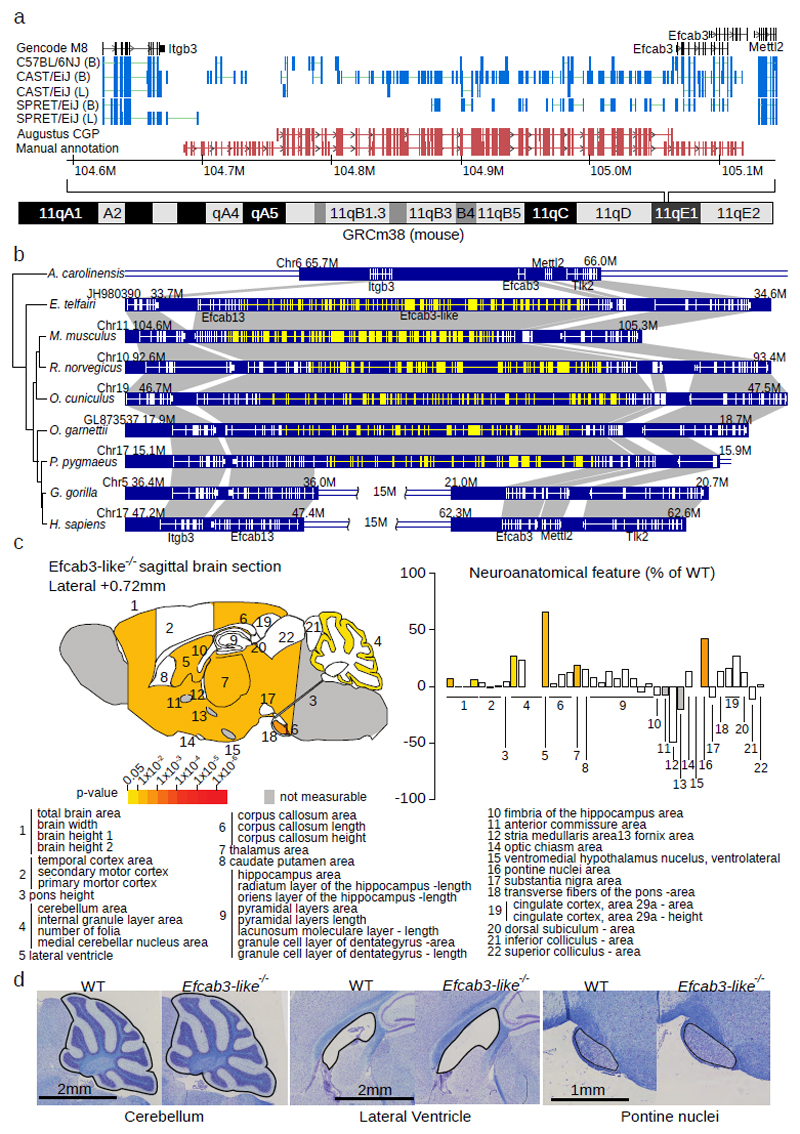
Figure 3. Efcab3-like locus, evolutionary history, and knockout phenotyping.
(a) Comparative Augustus identified an unannotated 188 exon gene (Efcab3-like, red tracks). RNA-Seq splicing from two tissues (B=Brain, L=Liver, blue tracks) and five strains are displayed. Manual annotation extended this gene to 188 exons (lower red track). (b) Evolutionary history of Efcab3-like in vertebrates including genome structure and surrounding genes. The mRNA structure of each gene is shown with white lines on the blue blocks. Novel coding sequence discovered in this study is shown in yellow. Notably, Efcab13 and Efcab3 are fragments of the novel gene Efcab3-like. A recombination event happened in the common ancestor of sub-family Homininae, which disrupted Efcab3-like in gorilla and chimpanzee human. (c) Schematic representation of 22 brain regions plotted in sagittal plane for Efcab3-like mutant male mice (16 weeks of age, n=3) according to p-values (two-tailed equal variance t-test, left). Corresponding brain regions are labelled with a number that is described below the panel (Supplementary Table 15). White colouring indicates a p-value > 0.05 and grey indicates that the brain region could not be confidently tested due to missing data. Histograms showing the neuroanatomical features as percentage increase or decrease of the assessed brain regions in Efcab3-like mutant mice compared to matched controls (right). (d) Representative sagittal brain images of matched controls (left) and Efcab3-like mutant (right), showing a larger cerebellum, enlarged lateral ventricle and increased size of the pontine nuclei (n=3, see Supplementary Figure 15).