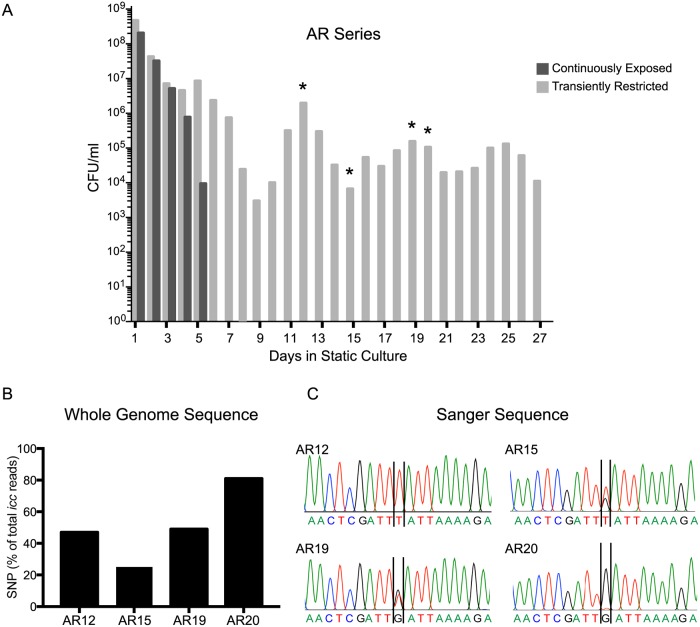
Fig 2. Independent validation of cyclical pattern of viability and microevolution.
(A) Viability was determined every 24 hours for 27 days. Asterisks denote cultures selected for whole genome sequencing in the AR series. (B) The sequence reads that exhibit the SNP are reported as percentage of the total reads for icc on each of the days. The number of sequence reads for each sample were: AR12 (726), AR15 (707), AR19 (1021) and AR20 (865). (C) Sanger sequencing electropherograms validate the mutation [Thymine (T) → Guanine (G)].