FIGURE 1.
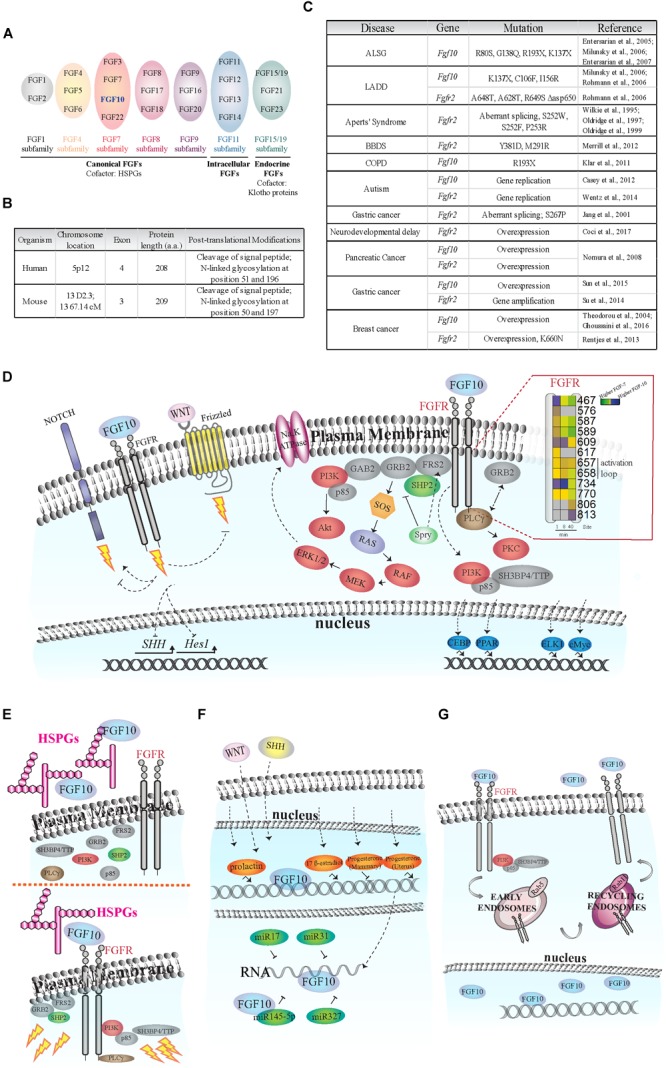
Fibroblast of Growth Factor (FGF) signaling activation and regulation. (A) Schematic of FGF subfamilies (see Ornitz and Itoh, 2015). FGF10 belongs to the FGF7 subfamily and is highlighted in blue. (B) Comparison of FGF10 gene/protein in human and mouse. The human FGF10 gene has an extra exon; and the protein length in the two species differs by one amino acid. FGF10 is secreted via the canonical ER-Golgi secretory pathway, as demonstrated by the cleavage of the signal peptide, and have two known glycosylation sites (Source: UniProt). (C) Known disease-causing mutations on Fgf10 and Fgfr2 genes. The rare developmental disorders Aplasia of Lacrimal and major Salivary Glands (ALSG), Lacrimo-Auricuo-Dentro-Digital (LADD) syndrome, Aperts’ Syndrome and Bent Bone Dysplasia Syndrome (BBDS) result from either the loss of key receptor-binding sites on FGF10 or mutations in the receptor kinase (TK) domains or IgII linker regions (Wilkie et al., 1995; Oldridge et al., 1997, 1999; Entesarian et al., 2005, 2007; Milunsky et al., 2006; Rohmann et al., 2006; Merrill et al., 2012). Interestingly, cases of ALSG caused by the R193X mutation also coincide with Chronic Obstructive Pulmonary Disease (COPD) (Klar et al., 2011). More recently, chromosomal translocation and duplication events at the loci of the Fgf10 and Fgfr2 genes have been associated with neurological disorders such as developmental delay and autism (Casey et al., 2012; Wentz et al., 2014). The role of single nucleotide polymorphisms and de novo point mutations causing oncogenic expression of Fgf10 and Fgfr2 are also becoming clearer, particularly in pancreatic, gastric, and breast cancers (Jang et al., 2001; Theodorou et al., 2004; Nomura et al., 2008; Reintjes et al., 2013; Su et al., 2014; Sun et al., 2015; Ghoussaini et al., 2016; Coci et al., 2017). (D) FGF10-dependent activation of FGFR intracellular tyrosine residues (Y; insert), adaptor proteins (gray), protein kinases (red), and transcription factors (blue). Insert on the right: each square represents a phosphorylated tyrosine residue (Y); the numbers correspond to phosphorylated Y residues on FGFR2b identified by proteomics; the color blue indicates higher phosphorylation at a given time point upon FGF10 stimulation of epithelial cells; modified from Francavilla et al. (2013). Dashed arrows represent FGF10-specific activation or inhibition of signaling. The lightning bolt represents the activation of signaling cascades. (E) Left, FGF10 bound to heparan sulfate proteoglycans (HSPGs) in the ECM does not activate FGFR and intracellular signaling. Right, FGF10 bound to HSPGs and FGFR induces the recruitment of protein adaptors to FGFR and signaling activation (represented by lightning bolts). (F) Schematic representation of FGF10 regulation in response to hormones (orange), miRNAs (dark green), WNT and SHH proteins. (G) FGF10 induces FGFR2b internalization into early endosomes and sorting to recycling endosomes and plasma membrane. FGF10 has also been found in the nucleus of certain cell types.
