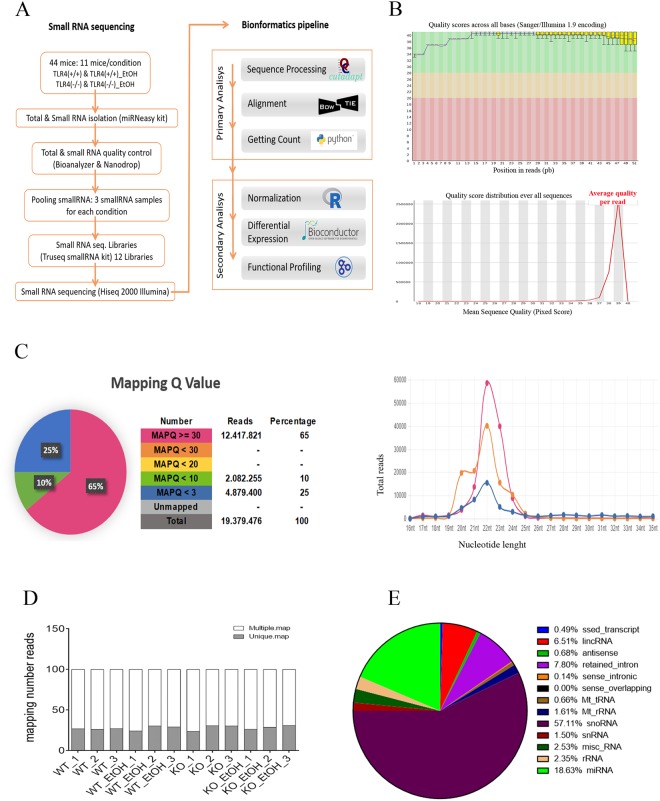
Figure 1.
Experimental NGS Workflow and small RNA Quality Control. Small RNA libraries were prepared from the cortices of 44 mice (11 mice/condition). Samples were used for the deep sequencing protocol on the Illumina platform. Bioinformatics primary and secondary pipelines were used to detect the miRNAs profile and the differential expression miRNAs analysis (A). The Quality Control score, the Q value in all the nucleotides sequenced, were higher than 39, and the length with the best Q values was 51 nucleotides (B). Mapping Quality of the reads and distribution MAPQ by nucleotide length (21–25nt represent miRNA reads) (C). Number of reads aligned in multiple loci or unique loci in the reference genome sequence (D). Percentage of reads mapping RNA species (E).