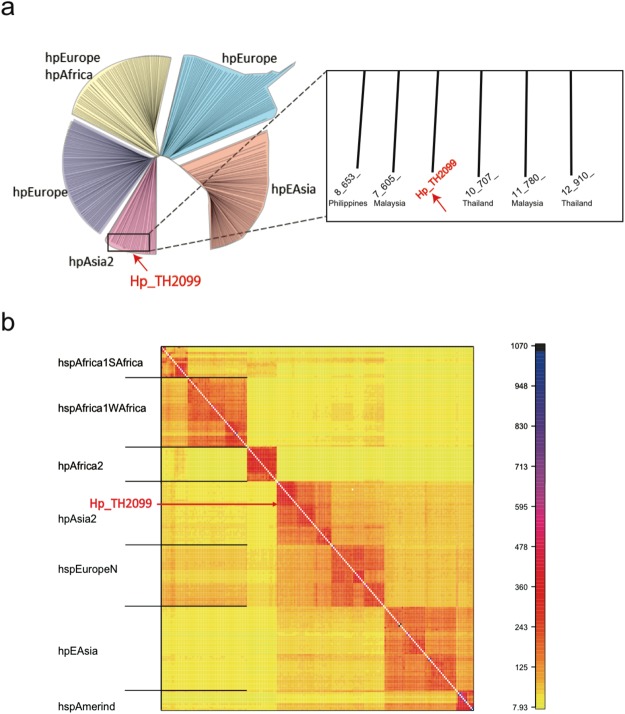
Figure 1.
Isolation of cagA-positive H. pylori from macaque stomach. (a) Molecular phylogenetic tree based on MLST analysis. Seven H. pylori genes (atpA, efp, mutY, ppa, trpC, urel, yphC) were used to perform MLST analysis (left). Magnified view of the phylogenetic tree in the vicinity of Hp_TH2099 (right). (b) fineSTRUCTURE analysis of the Hp_TH2099 genome. The color of each cell of the matrix indicates the expected number of DNA chunks imported from a donor genome (column) to a recipient genome (row). The boundaries between named populations are marked with lines. Detailed information on H. pylori isolates used in this analysis is shown in Supplementary Table S1.