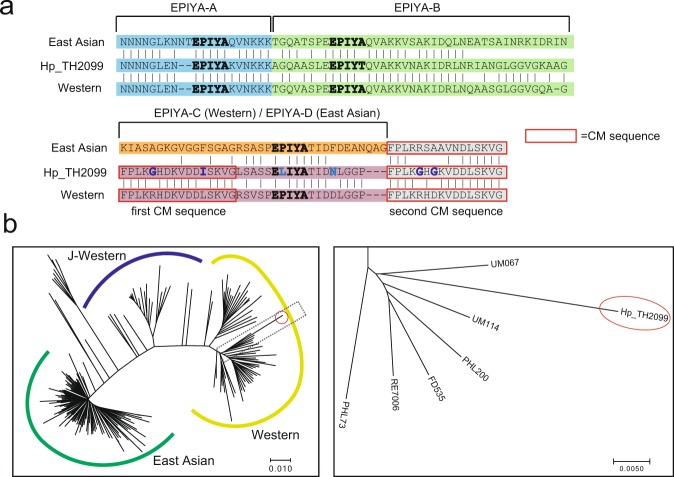
Figure 2.
Sequence analysis of Hp_TH2099 CagA. (a) Amino acid sequences for the EPIYA-repeat regions of canonical East Asian CagA, Hp_TH2099 CagA, and canonical Western CagA were aligned using Clustal W. Amino acid variations in Hp_TH2099 CagA are indicated in dark blue and light blue bold letters. (b) Phylogenetic tree based on the nucleotide sequence of the Hp_TH2099 cagA gene (shown in a red circle) with 199 full-length cagA genes extracted from the GenBank database (left). Positions of East Asian cagA, Western cagA and J-Western cagA were indicated in green line, yellow line, and blue line, respectively. Magnified view of the black dotted box in left figure (right). Hp_TH2099 and other closely related cagA genes were shown. The tree is drawn to scale with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distance was computed in the units of the number of base substitutions per site.