Abstract
Anthropogenic global warming, nitrogen addition, and overgrazing alter plant communities and threaten plant biodiversity, potentially impacting community productivity, especially in sensitive mountain grassland ecosystems. However, it still remains unknown whether the relationship between plant biodiversity and community productivity varies across different anthropogenic influences, and especially how changes in multiple biodiversity facets drive these impacts on productivity. Here, we measured different facets of biodiversity including functional and phylogenetic richness and evenness in mountain grasslands along an environmental gradient of elevation in Yulong Mountain, Yunnan, China. We combined biodiversity metrics in a series of linear mixed‐effect models to determine the most parsimonious predictors for productivity, which was estimated by aboveground biomass in community. We examined how biodiversity–productivity relationships were affected by experimental warming, nitrogen addition, and livestock‐grazing. Species richness, phylogenetic diversity, and single functional traits (leaf nitrogen content, mg/g) represented the most parsimonious combination in these scenarios, supporting a consensus that single‐biodiversity metrics alone cannot fully explain ecosystem function. The biodiversity–productivity relationships were positive and strong, but the effects of treatment on biodiversity–productivity relationship were negligible. Our findings indicate that the strong biodiversity–productivity relationships are consistent in various anthropogenic drivers of environmental change.
Keywords: experimental warming, functional diversity, grazing, multimodel inference, nitrogen addition, phylogenetic diversity
1. INTRODUCTION
Anthropogenic impacts such as increasing temperature, higher nitrogen addition, and overgrazing all conspire to cause rapid declines in plant biodiversity worldwide, especially in mountain grassland ecosystems, which naturally elicits concern about the consequences for the maintenance of ecosystem functioning (Chapin et al., 2000; Cingolani, Noy‐Meir, & Diaz, 2005; Roth, Kohli, Rihm, & Achermann, 2013; Urban, 2015). The relationship between plant biodiversity and ecosystem function has been a major research topic in ecology for several decades, and while there is general empirical support for a positive effect of biodiversity on function from manipulative experiments (Balvanera et al., 2006; Cardinale et al., 2006; Tilman, Isbell, & Cowles, 2014), there is a lack of clarity about how anthropogenic changes in plant biodiversity might affect biomass production in more natural systems (Zavaleta & Hulvey, 2007). Inconsistent biodiversity effects on productivity could result from how biodiversity is measured, the confounding effect of environmental heterogeneity, and the nature of the anthropogenic impacts.
One reason might be that traditional biodiversity measures, like species richness, do not sufficiently capture the critical processes such as resource complementarity and interspecific interactions that are responsible for ecosystem function, which might be better reflected in relevant traits or evolutionary histories of species in a community (Lavorel & Garnier, 2002; Partel, Laanisto, & Zobel, 2007). Recently, a number of studies have shown that measures based on phylogenetic or single or multiple functional traits appear to be superior to species richness in explaining variation in productivity of plant communities (Cadotte, 2013; Cadotte, Cavender‐Bares, Tilman, & Oakley, 2009; Flynn, Mirotchnick, Jain, Palmer, & Naeem, 2011; Liu, Zhang et al., 2015), and further supply direct links to the mechanisms controlling productivity (Cadotte, 2017). In addition to this, some studies (Liu, Zhang et al., 2015) found that statistical models that combined different biodiversity facets maximally explained the effects of biodiversity loss on ecosystem functioning or services. For example, Liu, Zhang et al. (2015) found that multivariate functional diversity was the single predictor that consistently outperformed other single‐biodiversity measures in explaining variation in productivity, but phylogenetic diversity and community‐level plant height combined to explain maximum variation. However, beyond biodiversity facets that represent species‐level differences, intraspecific variation is critically important to fully capture the diversity of plant communities (Albert et al., 2012). Ali and Mattsson (2017) evaluated the relative power of intraspecific and interspecific tree size variation and found that intraspecific variation better explained variation in aboveground biomass.
Although biodiversity is a major determinant of ecosystem productivity, the estimation of the biodiversity effect might be confounded by environmental factors and potential drivers of environmental change such as elevated temperature, nitrogen addition, and herbivory (Fridley, 2002; Hooper et al., 2005; Seabloom et al., 2017; Steudel et al., 2012; Tilman, Reich, & Isbell, 2012; Tilman et al., 2014). Thus, it is important to disentangle the relative importance of biodiversity relative to other drivers along an environmental gradient for inferring the consistent effects of biodiversity on the primary productivity of ecosystems. The majority of research on biodiversity effects on ecosystem function has been in experimentally assembled communities, and these studies generally support a positive relationship between biodiversity and ecosystem functioning (Hector et al., 1999; Liu, Zhang et al., 2015; Tilman et al., 2001). In contrast, biodiversity levels produced by an environmental gradient such as elevation might reveal different response of ecosystem productivity (Gough, Grace, & Taylor, 1994). Hence, the direct relevance of these experiments for estimating the impacts of realistic biodiversity loss due to environmental changes on ecosystem functioning remains controversial (Hector et al., 2007; Jiang, Wan, & Li, 2009).
To address the biodiversity–productivity relationships of natural communities under different anthropogenic impacts, we developed a fenced warming‐fertilizing experiment in mountain wetlands along an elevation gradient on Yulong Mountain, Yunnan, China. We employed open‐topped, passive warming chambers and urea fertilizer to simulate the projected global warming and nitrogen addition, respectively. We used a multimodel comparative approach to assess the relative contribution of single and various combinations of multivariate biodiversity indices, both with and without intraspecific variation, to predict the variance in biomass production after accounting for potential confounding factors including local environmental heterogeneity, warming, fertilizing, and grazing. We aimed to answer the following questions: (a) Does phylogenetic and functional diversity outperform traditional richness and evenness regardless of environmental heterogeneity and anthropogenic impacts? (b) Does incorporating intraspecific trait variability enhance the explanatory power of functional diversity? (c) Are biodiversity–productivity relationships comparable in experimental warming, nitrogen addition, and grazing along environmental gradient of elevation in mountain grasslands?
2. MATERIALS AND METHODS
2.1. Study sites and experimental design
We established eighteen study sites in south‐facing wetlands of regular topology of Yulong Mountain (100°10′E, 27°00′N) along an environmental gradient of elevation (2,700, 3,200, and 3,400m) within the Lijiang Alpine Botanical Garden of the Kunming Institute of Botany, Chinese Academy of Sciences in Lijiang, Yunnan Province, China. Yulong Mountain has the mean annual temperature of 12.8°C and the annual rainfall is 935 mm, which is mainly distributed from July to October with distinct dry and rainy seasons (Luo et al., 2016). Plant communities of wetlands have obvious species turnover along the elevation gradient with the dominance of the genera Isachne, Juncus at the lowest elevation, the genera Ligularia, Agrostis at the middle elevation, and the genus Agrostis at the highest elevation. All three wetlands have long livestock‐grazing histories, and each supports different types of livestock where sheep and horses graze at the lowest elevation, scalpers, and yaks graze at the middle and highest elevation, respectively.
We established six 12 × 12 m permanent fenced sites randomly distributed in wetlands within each elevation in May 2015 (Figure 1). Within each permanent site, we conducted a complete randomized block factorial experiment with each block of size 5 × 5 m. There were two factors of both experimental warming and nitrogen addition in each block and two levels for each factor. In both fertilized blocks, we applied urea fertilizer annually at the beginning of the rainy season approximately the end of May at a rate of 5 g m−2 year−1. In both warmed blocks, we applied open top chambers (OTCs), commonly employed devices to study the effects of climate warming on ecosystems (Marion et al., 1997). Here, our open top chambers were octahedral frames made of angle iron, 1.5 m maximum diameter, and 45 cm height. Six sides of each open top chamber were fastened to transparent 1.5‐mm‐thick hard plastic with adjacent edges of two plastic pieces attached with adhesive. We regularly arranged two open top chambers and two corresponding plots of the similar area in four blocks with at least 3 m between the nearest edges of adjacent plots. Furthermore, we randomly positioned 3–4 plots around each permanent site with total of 20 grazed plots. Hence, there were five treatments (T C = control, T W = warming, T N = nitrogen addition, T WN = combination of warming and nitrogen addition, and T G = livestock‐grazing; Figure 1).
Figure 1.
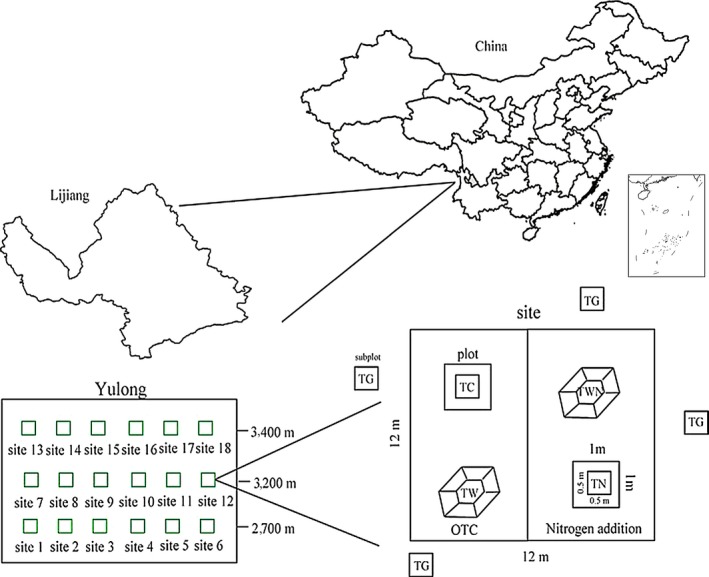
Map of the study sites on Yulong Mountain, Lijiang, Yunnan Province, China, and the plot design. Shown are treatments: T C = control, T W = warming, T N = nitrogen addition, T WN = warming and nitrogen addition, and T G = livestock‐grazing
We recorded species richness and their abundance in a rectangular subplot of 0.5 × 0.5 m from the center of each plot at the peak of the growing season in August 2016 (Figure 1). We then harvested all the stems of each species in each subplot at ground level, dried, and weighed them to 0.1 mg to estimate biomass production (productivity).
2.2. Environmental data
After cutting the stems to ground level, we collected soil core samples from three random locations in each subplot with a cylindrical soil auger (5 cm inner diameter, 15 cm length). We combined the three replicates from the same depth for each subplot as a single composite sample, dried it in the shade, and filtered it using a 2‐mm sieve for stoichiometric analysis. We measured soil pH, concentration of nitrogen (N), phosphorus (P), and carbon (C) following the standard protocols (Sparks et al., 1996). Besides soil resources, we also collected climatic data for rainfall, air temperature, and air moisture using HOBO RG3‐M, HOBO Pro v2, respectively (Onset Computer Corporation, Bourne, MA, USA) from July to October in 2016. For each elevation, we placed one HOBO RG3‐M and two HOBO Pro v2, of which one was positioned inside an open top chamber and the other one was positioned in a control plot. We showed the detailed distributions of temperature and moisture during the experimental interval in Figure A1.
2.3. Plant traits and community phylogeny
We measured five plant traits including plant height (H, cm), leaf carbon content (LC, mg/g), leaf nitrogen content (LN, mg/g), leaf phosphorus content (LP, mg/g), and specific leaf area (SLA, cm2/g). These plant traits might reflect fundamental resource complementarity and interactions among co‐occurring species (Weiher et al., 1999; Wright et al., 2004). We recorded plant height of maximum five randomly selected individuals from each species in each subplot. We calculated the maximum of plant height for each species per plot for intraspecific variability among plots. We scanned at least 1 mature leaf of randomly selected five individuals per species in each subplot using an Epson‐V200 scanner. We then measured leaf area with image analysis software (ImageJ; http://rsb.info.nih.gov/ij). We weighed the leaves after dried to a constant weight at 60°C to 0.1 mg and calculated the specific leaf area as the ratio of leaf dry mass to leaf area. We pooled the leaves from different individuals of the same species and measured leaf carbon, nitrogen, and phosphorus content. For the missing traits data due to rare species, we substitute the average of the same traits of the same species or same genus within the same treatment.
We constructed the phylogeny for the 105 species recorded in our study using rbcL + matK regions of the chloroplast genome. The detailed descriptions of DNA extraction, amplification, and sequencing are provided in Liu, Yan et al. (2015). Here, we briefly described the inference method of phylogenetic reconstruction. We aligned the rbcL and matK sequences using MAFFT (Katoh & Standley, 2013) and concatenated matK to the rbcL to form a super matrix. We used the sequences from the same genus in BOLD as the substitutes for the missing sequences in 27 of the species. For each gene, we selected top‐ranked maximum‐likelihood model of nucleotide substitution using Akaike's information criterion, as implemented in the function modelTest in the phangorn library (Schliep, 2011) in R (R Core Team, 2016). Then, we estimated a maximum‐likelihood phylogeny using PhyML 3.0 with the starting‐tree estimated from the BioNJ (Guindon et al., 2010). We chose one representative of early diverging angiosperm lineage Amborella trichopoda as the root of phylogeny and then used a semiparametric rate‐smoothing method to transform the phylogeny to an ultrametric tree using the chronopl function with parameter value 1,000 in the R ape library (Paradis, Claude, & Strimmer, 2004).
2.4. Measures of biodiversity
Using species composition and number of individuals, we calculated traditional species richness (S) and Shannon's evenness index (H′) for each subplot. We also calculated a suite of single and multivariate functional diversity metrics based on plant traits, as well as phylogenetic diversity metrics using the maximum‐likelihood phylogeny. We listed the detailed descriptions of the measures of biodiversity in Table A1. Here, we give a brief description of important functional and phylogenetic metrics. To assess the potential effect of intraspecific trait variability, we averaged the traits for each species across all subplots in the study as its “fixed” traits and averaged the traits for each species in a given subplot as its “specific” traits. We then calculated a number of functional diversity metrics including single community‐level plant traits and multivariate functional diversity metrics for each subplot using both “fixed” and “specific” traits (Leps, de Bello, Smilauer, & Dolezal, 2011). Here, multivariate functional diversity metrics included Rao's quadratic entropy (RaoQ), which measures abundance‐weighted distances based on multiple traits (Botta‐Dukat, 2005) and functional richness (FRic), which measures the volume of the functional space occupied by the community (Villeger, Mason, & Mouillot, 2008). For the measures of phylogenetic diversity, we calculated the imbalance of abundances at higher clades (IAC), which encapsulates the distribution of individuals across the nodes in the phylogeny (Cadotte et al., 2010) and the abundance‐weighted mean nearest taxon distance in an assemblage (MNND; Cadotte et al., 2010).
2.5. General linear mixed‐effect models
We constructed a series of general linear mixed‐effect models to determine the most parsimonious relationships between productivity and the various measures of biodiversity, treatment, and local environmental factors including soil resources. We assumed that various measures of biodiversity, experimental treatments, and soil resources as fixed factors, whereas elevation, treatment, and plot were treated as hierarchical random factors. Here, the use of a normal distribution of model residuals was validated based on the normalized scores of standardized residual deviance (Q–Q plots). To evaluate model support, we used Akaike's information criterion corrected for small sample sizes (AICc; Burnham & Anderson, 2002, 2004). We also used the marginal R 2 values of the models (R m 2) as a measure of the model's goodness of fit (Nakagawa & Schielzeth, 2013).
To search for the most parsimonious models explaining patterns of biomass production, we firstly removed redundant predictors associated with phylogenetic, functional diversity metrics. We selected the relatively better‐ranked single‐biodiversity metric models in both phylogenetic and functional diversity metrics. Meanwhile, to testify whether experimental treatments affect biodiversity–productivity relationships, we regressed biomass production against each biodiversity metric with the addition or multiplication of treatment and compared the explanatory ability of these models using Akaike's information criterion weights. The detailed single‐biodiversity model ranking is listed in Table A2 and the biodiversity metrics we used in the following model construction are listed in Table 1.
Table 1.
Measures of biodiversity for general multivariate linear mixed‐effect models
| Biodiversity measure | Description | References |
|---|---|---|
| IAC | Imbalance of abundances among clades: measures the deviation in abundance distribution among internal splits from a null | Cadotte et al. (2010) |
| H max | Community‐level mean of plot‐specific maximum plant height values | Leps et al. (2011) |
| LN | Community‐level mean of mean leaf nitrogen content value for individual species used for all plots where the species is found | Leps et al. (2011) |
| S | Realized species richness of plot | Tilman, Wedin, and Knops (1996) |
| MNND | Mean nearest neighbor distance (the mean of the shortest distances connecting each species to any other species in the assemblage) | Webb, Ackerly, McPeek, and Donoghue (2002) |
| RaoQ | Quadratic entropy using plot‐specific trait values | Botta‐Dukat (2005) |
| FDis | Functional dispersion: weighted distances from a weighted centroid in multitrait space using plot‐specific trait values | Villeger et al. (2008) |
| H′ | Shannon's diversity index | Tilman et al. (1996) |
The order from top to bottom for the measures of biodiversity represents their relative ranking using Akaike's information criterion weights.
Because of the strong correlation between most biodiversity indices (Spearman's ρ > 0.3; Table A3) and because multivariate functional indices are derived from the same trait data, we avoided including more than one of these like indices in any one model. Then, we constructed models with all remaining combinations of selected biodiversity metrics. At the meanwhile, we incorporated the interaction term between selected biodiversity metrics and experimental treatment into the model if multipliable model outperformed additive model considering treatment effects for particular selected biodiversity metrics. Finally, we also incorporated soil resources into above constructed models following the same constraint of correlation among soil resources and between selected biodiversity metrics and soil resources.
3. RESULTS
3.1. Comparisons between biodiversity metrics
As expected, phylogenetic and functional diversity indices alone outperformed traditional species richness and Shannon's evenness to explain the variation of biomass production when simultaneously considering elevation and treatment (Table 1, Table 2, Tables A2 and A4). Although only several functional diversity indices (H max, RaoQ, FDis, FDiv, detailed information see in Table A1) considering intraspecific variability attained greater model support than corresponding indices using species mean traits (Table 1, Table A2), most of these indices were selected as relatively better‐ranked single‐biodiversity metrics (Table 1). Of all functional diversity indices, the community‐level mean of “specific” maximum plant height (H max) on average accounted for the most explained variation in biomass production (R m 2 > 50%; Table 1). Phylogenetic diversity (IAC) was the top‐ranked single‐biodiversity metric of all considered biodiversity metrics here and explained the most variation in biomass production (R m 2 > 66%; Table A2).
Table 2.
General linear mixed‐effect model (GLMM) results for biomass production as a function of several fixed factors and a hierarchical random factor
| Model | LL | k | AICc | ΔAICc | wAICc | R m 2 | R c 2 |
|---|---|---|---|---|---|---|---|
| S + IAC + LN + C + T | 57.115 | 13 | −85.145 | 0.000 | 0.291 | 91.8 | 99.9 |
| S + IAC + LN + T | 55.664 | 12 | −84.706 | 0.439 | 0.234 | 91.6 | 99.9 |
| S + IAC + LN + N + T | 56.336 | 13 | −83.588 | 1.557 | 0.134 | 91.7 | 99.9 |
| S + IAC + C + T | 54.529 | 12 | −82.437 | 2.708 | 0.075 | 91.4 | 99.9 |
| S + IAC + T | 53.195 | 11 | −82.189 | 2.955 | 0.066 | 91.3 | 99.9 |
| S + IAC + FDis + C + T | 55.010 | 13 | −80.934 | 4.210 | 0.035 | 91.5 | 99.9 |
| S + IAC + N + T | 53.732 | 12 | −80.843 | 4.302 | 0.034 | 91.3 | 99.9 |
| S + IAC + FDis + T | 53.725 | 12 | −80.829 | 4.316 | 0.034 | 91.3 | 99.9 |
| S + IAC + RaoQ + C + T | 54.906 | 13 | −80.726 | 4.418 | 0.032 | 91.5 | 99.9 |
| S + IAC + RaoQ + T | 53.642 | 12 | −80.661 | 4.483 | 0.031 | 91.3 | 99.9 |
Fixed factors are number of species (S), Shannon's evenness (H’), and phylogenetic diversity (IAC, imbalance of abundance at the clade; MNND, mean nearest‐neighbor distance), and community‐level mean of single functional traits (H max, plot‐specific maximum plant height; LN, mean leaf nitrogen content value for individual species used for all plots where the species is found) or multivariate functional trait indices (RaoQ, Quadratic entropy; FDis, Functional dispersion: weighted distances from a weighted centroid in multitrait space), and experimental treatments (T: T C = control, T W = warming, T N = nitrogen addition, T WN = warming and nitrogen addition and T G = livestock‐grazing), and soil resources (C, soil carbon content; N, soil total nitrogen content; P, soil total phosphorus content). Hierarchical random factor is elevation (2,700, 3,200, and 3,400 m), treatment, and plot. Values are shown for the estimated number of model parameters (k), maximum log‐likelihood (LL), and the information‐theoretic Akaike's information criterion corrected for small samples (AICc), change in AICc relative to the top‐ranked model (ΔAICc), AICc weight (wAICc, model probability), and the marginal and total variance explained (R m 2, R c 2) as a measure of the model's goodness of fit. The top 10 models are listed; the full table is shown in Appendix: Table A3.
3.2. Biodiversity effects
Of the 166 multivariate linear mixed‐effect models, the most parsimonious model included species richness (S), phylogenetic diversity (IAC), the community‐level mean of “fixed” leaf nitrogen content (LNf), soil carbon content (C), and treatment (T) accounting for >91% of the deviance explained in productivity (Table 2). After accounting for confounding effects of environmental factors and experimental treatment, biomass production generally increased with increasing species richness, phylogenetic diversity, and the community‐level mean of “fixed” leaf nitrogen content (Figure 2a–c).
Figure 2.
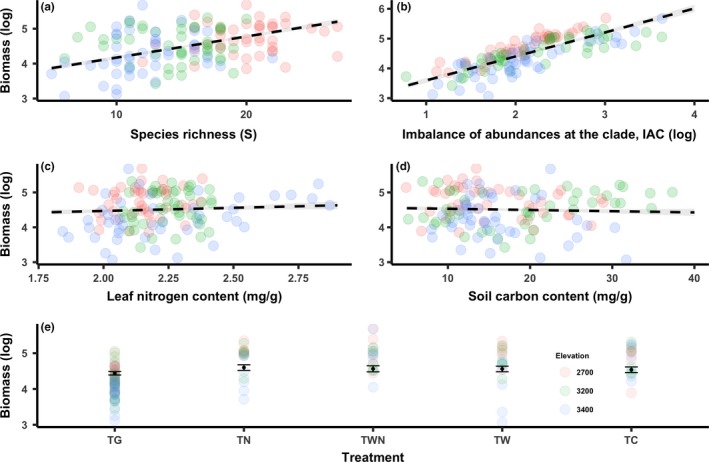
Scatter plots of the best‐supported variables combined in the general linear mixed‐effect models to predict variation in biomass production: (a) species richness (S), (b) imbalance of abundance at the clade (IAC) based on a maximum‐likelihood phylogeny, (c) community‐level mean of mean leaf nitrogen content value for individual species used for all plots where the species is found (LN), (d) soil carbon content (C), and (e) experimental treatments (T C = control, T W = warming, T N = nitrogen addition, T WN = warming and nitrogen addition, and T G = livestock‐grazing). Dashed lines are linear regression lines, gray ribbon are their confidence intervals, and points and error bar in (e) are predicted values and their confidence intervals using general linear mixed‐effect model
3.3. Environmental and treatment effects
We found relatively weaker environment and treatment effects on biomass production compared to those of selected biodiversity metrics, but few treatment effects on biodiversity–production relationship (Table 2, Table A2). The top‐ranked model showed that grazing strongly reduced the biomass production compared with nitrogen addition; however, nitrogen addition and experimental warming showed no impact on biomass production (Figure 2e). We also found evidence for a weak negative relationship between biomass production and soil carbon content (Figure 2d).
4. DISCUSSION
Our results show that phylogenetic and functional diversity alone outperformed traditional biodiversity measures, species richness, and Shannon's evenness, for explaining variation in productivity. This corroborates observational and experimental evidence that phylogenetic and functional measures better align with the mechanisms controlling community assembly and ecosystem function than taxonomic measures (Cadotte et al., 2009; Flynn et al., 2011; Liu, Zhang et al., 2015). Of all considered functional biodiversity indices, a single functional trait was the single best predictor of productivity patterns. This is not surprising since single functional trait might explain a larger amount of variation in productivity than multivariate functional indices likely due to functional trade‐offs and coordinated variation of functional traits (Cingolani, Cabido, Gurvich, Renison, & Diaz, 2007; Roscher et al., 2012).
Meanwhile, our study revealed that transitioning from using species mean (e.g., “fixed”) traits to plot level (e.g., “specific”) traits enhanced the explanatory power of functional diversity irrespective of plant traits in isolation or combination. Including specific traits allows us to detect subtle differences in functional diversity that respond to environmental variation that does not involve species turnover (Luo et al., 2016). Indeed, Jung et al. (2014) reported that the response of subalpine grassland communities to short extreme drought events was more mediated by intraspecific trait variability than species turnover. Intraspecific trait variability, through phenotypic plasticity, can promote species coexistence through providing fitness advantages and acting as a buffer against rapid climate change (Aspinwall et al., 2015; Nicotra et al., 2010; Valladares, Gianoli, & Gomez, 2007). This might lead to the shift in plant strategies in association with resource capture and use efficiencies at the local scale, which in turn are more related to plot‐specific aboveground biomass production. Furthermore, phenotypic plasticity, especially associating with maximum plant height, might ameliorate light competition, which is assumed to be an important mechanism explaining species loss and biodiversity effects (Borer et al., 2014; Cadotte, 2017; Fridley, 2003; Hautier, Niklaus, & Hector, 2009; Zhou et al., 2017). Our results generally supported these assumptions and highlighted the critical role of intraspecific trait variability in more precisely predicting the ecosystem functioning in the face of global climate change.
Although functional diversity could explain a substantial proportion of variation in productivity, the combination of phylogenetic diversity and a functional trait (leaf nitrogen) attained more model support and greater explanatory power. This implies that functional diversity and phylogenetic diversity could complement each other in the perspective of ecosystem functioning because of their own limitations. Functional diversity was limited by the absence of potential key functional traits, for example, belowground root traits in our study (Cadotte et al., 2009). Linkage between phylogenetic diversity and real ecological differences remains unclear (Cadotte, Davies, & Peres‐Neto, 2017). Thus, the influence of unmeasured plant traits might be compensated by metrics that capture phylogenetic information, such as the distribution of abundances at the clades or the equitability of abundance‐weighted entropic measure of the distribution of evolutionary distinctiveness in an assemblage (Cadotte et al., 2010). Such a combination of functional and phylogenetic information for explaining biodiversity–productivity relationships has received support from both biodiversity manipulation experiments and natural ecosystems (Liu, Zhang et al., 2015; Zhou et al., 2017). For example, Liu, Zhang et al. (2015) found that phylogenetic diversity and plant height represented the most parsimonious combination to predict aboveground biomass production in a removal experiment where species richness and functional diversity were manipulated in alpine meadows of the Tibetan Plateau.
In this study, we found strong and positive effects of species richness on productivity in natural ecosystems after accounting for potential confounding factors. This was consistent with a review by Tilman et al. (2014), in which the diversity effect is as great as, or greater than, the effects of herbivory, nitrogen addition, and other drivers of environmental change. Although our experiment is limited in the short term by the drivers of environmental change, our results still supported a strong positive species richness–productivity relationship in natural ecosystems even after quantifying the effects of intraspecific trait variability and evolutionary history. Despite our findings, the role of biodiversity in the productivity of natural ecosystem remains controversial (Adler et al., 2011), and our results emphasize the fact that we underestimate the importance of biodiversity for ecosystem function when we use species richness only.
Our results revealed that the drivers of environmental change had negligible effects on the relationship between biodiversity and aboveground biomass production. Our finding showed that the relationship between IAC and biomass production was consistently strongest for all considered biodiversity metrics in various treatments. Cadotte (2013) showed that biomass production was strongly predicted by phylogenetic diversity and that this finding might result from species complementarity, and ultimately species coexistence mechanisms (Chesson & Warner, 1981; Hodapp, Hillebrand, Blasius, & Ryabov, 2016; Horn & Macarthur, 1972; Levins & Culver, 1971). IAC that quantifies the relative deviation in the abundance distribution of a local community from a null distribution where individuals are evenly partitioned between clade splits can be used to infer the relative importance of competition and environmental filtering for local assembly. IAC would tend toward 0 if the strength of competition was proportional to phylogenetic relatedness, while IAC would be far greater than 0 if environmental filtering was key to community structure (Cadotte et al., 2010). Meanwhile, Cadotte (2017) showed that multidimensional trait measures might drive complementarity effect through niche complementarity, while few, singular traits (mainly height) might drive selection effect through interspecific competition. Our results were generally in line with these studies, because on the one hand, maximum plant height outperformed the multivariate functional indices alone in the perspective of ecosystem productivity, implying the importance of selection effect in biomass production in natural mountain grassland ecosystems; on the other hand, we observed IAC values far greater than 0, implying the dominance of environmental filtering in local community assembly, which might contribute to the role of selection effect in our system. Our results point to the importance of both complementarity effects and selection effects for aboveground biomass production in natural mountain grassland ecosystems.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
JJL conceived the idea and designed the experiment, JJL collected the data, JJL and MWC analyzed the data, JJL and MWC led the writing of the manuscript, and all authors contributed critically to the drafts and gave final approval for publication.
DATA ACCESSIBILITY
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.5b02c11
ACKNOWLEDGEMENTS
This research was funded by the National Natural Science Foundation of China (31500335, 31570210) and Ministry of Science and Technology of the People’s Republic of China (2014CB954100).
APPENDIX 1.
1.1.
Table A1.
Measures of biodiversity considered in the analysis
| Diversity measure | Description | References |
|---|---|---|
| S | Realized species richness of plot | Tilman et al. (1996) |
| H′ | Shannon's diversity index | Tilman et al. (1996) |
| PD | Phylogenetic diversity (the sum of all phylogenetic branch lengths connecting species together) | Faith (1992) |
| MNND | Mean nearest neighbor distance (the mean of the shortest distances connecting each species to any other species in the assemblage) | Webb et al. (2002) |
| MPD | Mean pairwise distance (the mean of all the distances connecting species in an assemblage) | Webb et al. (2002) |
| MNNDed | Abundance‐weighted MNND | Webb et al. (2002) |
| MPDed | Abundance‐weighted MPD | Webb et al. (2002) |
| EAED | Equitability of abundance‐weighted entropic measure of the distribution of evolutionary distinctiveness in an assemblage | Cadotte et al. (2010) |
| IAC | Imbalance of abundances among clades: measures the deviation in abundance distribution among internal splits from a null | Cadotte et al. (2010) |
| FEve | Functional Evenness: abundance‐weighted pairwise functional distances | Villeger et al. (2008) |
| FDis | Functional dispersion: weighted distances from a weighted centroid in multitrait space | Villeger et al. (2008) |
| FDiv | Functional divergence: mean abundance‐weighted deviance from an absolute abundance‐weighted deviance | Villeger et al. (2008) |
| FRic | Functional richness: convex hull volume of the trait space | Cornwell, Schwilk, & Ackerly, 2006; Villeger et al. (2008) |
| RaoQ | Quadratic entropy | Botta‐Dukat (2005) |
Figure A1.

(a) Cumulative rainfall, (b) mean air temperature within the open top chamber (OTC) and control subplots, and (c) mean moisture within the open top chamber (OTC) and control subplots at the field sites (elevation: 2,700, 3,200, and 3,400) in the growing season (July–October) in 2016
Table A2.
General linear mixed‐effect model (GLMM) results for biomass production as a function of single‐biodiversity metric, experimental treatment, and their interaction as fixed factor and hierarchical random factor
| Model | LL | k | AICc | ΔAICc | wAICc | R m 2 | R c 2 |
|---|---|---|---|---|---|---|---|
| IAC + T | −8.457 | 10 | 38.732 | 0.000 | 0.948 | 66.7 | 97.9 |
| IAC × T | −6.473 | 14 | 44.535 | 5.803 | 0.052 | 67.7 | 99.9 |
| H maxs + T | −44.775 | 10 | 111.367 | 72.635 | <0.001 | 50.2 | 98.2 |
| H maxs × T | −44.220 | 14 | 120.029 | 81.297 | <0.001 | 50.4 | 99.9 |
| LNf + T | −63.666 | 10 | 149.151 | 110.419 | <0.001 | 38.3 | 99.9 |
| S + T | −63.748 | 10 | 149.315 | 110.583 | <0.001 | 46.8 | 99.9 |
| S × T | −58.939 | 14 | 149.468 | 110.735 | <0.001 | 49.3 | 99.9 |
| LNs + T | −64.135 | 10 | 150.088 | 111.356 | <0.001 | 37.8 | 99.9 |
| NND + T | −65.273 | 10 | 152.364 | 113.632 | <0.001 | 41.1 | 99.9 |
| SLAf + T | −65.969 | 10 | 153.755 | 115.023 | <0.001 | 37.3 | 99.9 |
| PD + T | −66.131 | 10 | 154.081 | 115.349 | <0.001 | 43.4 | 99.9 |
| PD × T | −61.507 | 14 | 154.603 | 115.871 | <0.001 | 46.1 | 99.9 |
| HEED + T | −67.131 | 10 | 156.080 | 117.348 | <0.001 | 36.4 | 99.9 |
| LNf × T | −62.699 | 14 | 156.988 | 118.256 | <0.001 | 38.9 | 99.9 |
| LNs × T | −62.708 | 14 | 157.006 | 118.274 | <0.001 | 38.7 | 99.9 |
| NRIed + T | −67.609 | 10 | 157.037 | 118.305 | <0.001 | 35.2 | 99.9 |
| RaoQs + T | −67.614 | 10 | 157.047 | 118.315 | <0.001 | 36.3 | 99.9 |
| SLAs × T | −62.735 | 14 | 157.059 | 118.327 | <0.001 | 42.1 | 99.9 |
| FDiss + T | −67.792 | 10 | 157.403 | 118.670 | <0.001 | 36.5 | 99.9 |
| LPs + T | −67.829 | 10 | 157.477 | 118.744 | <0.001 | 33.3 | 99.9 |
| LPf + T | −67.870 | 10 | 157.557 | 118.825 | <0.001 | 33.3 | 99.9 |
| H′ × T | −63.169 | 14 | 157.927 | 119.195 | <0.001 | 40.1 | 99.9 |
| MPD + T | −68.332 | 10 | 158.483 | 119.751 | <0.001 | 35.1 | 99.9 |
| NNDed + T | −68.340 | 10 | 158.498 | 119.765 | <0.001 | 36.5 | 99.9 |
| SLAf × T | −63.507 | 14 | 158.604 | 119.872 | <0.001 | 39.1 | 99.9 |
| NND × T | −63.713 | 14 | 159.016 | 120.284 | <0.001 | 41.9 | 99.9 |
| H′ + T | −68.852 | 10 | 159.523 | 120.79 | <0.001 | 36.1 | 99.9 |
| LCf + T | −68.908 | 10 | 159.635 | 120.903 | <0.001 | 35.1 | 99.9 |
| MPDed × T | −64.030 | 14 | 159.650 | 120.918 | <0.001 | 39.0 | 99.9 |
| SLAs + T | −69.078 | 10 | 159.975 | 121.243 | <0.001 | 35.2 | 99.9 |
| FDisf + T | −69.096 | 10 | 160.010 | 121.278 | <0.001 | 35.7 | 99.9 |
| RaoQf + T | −69.164 | 10 | 160.146 | 121.414 | <0.001 | 35.5 | 99.9 |
| FRicf + T | −69.264 | 10 | 160.347 | 121.615 | <0.001 | 38.5 | 99.9 |
| NRI + T | −69.288 | 10 | 160.393 | 121.661 | <0.001 | 34.3 | 99.9 |
| NTI + T | −69.311 | 10 | 160.441 | 121.709 | <0.001 | 35.2 | 99.9 |
| NTIed + T | −69.648 | 10 | 161.115 | 122.383 | <0.001 | 34.7 | 99.9 |
| FDivs + T | −69.710 | 10 | 161.238 | 122.506 | <0.001 | 35.2 | 99.9 |
| LPs × T | −64.965 | 14 | 161.520 | 122.788 | <0.001 | 35.1 | 99.9 |
| LCs + T | −69.879 | 10 | 161.576 | 122.844 | <0.001 | 34.7 | 99.9 |
| LPf × T | −65.036 | 14 | 161.662 | 122.930 | <0.001 | 35.1 | 99.9 |
| H maxf + T | −70.006 | 10 | 161.830 | 123.098 | <0.001 | 33.9 | 99.9 |
| FRics × T | −65.158 | 14 | 161.906 | 123.173 | <0.001 | 39.5 | 99.9 |
| FDiss × T | −65.229 | 14 | 162.047 | 123.315 | <0.001 | 38.7 | 99.9 |
| LCf × T | −65.234 | 14 | 162.058 | 123.326 | <0.001 | 38.5 | 99.9 |
| NRIed × T | −65.310 | 14 | 162.210 | 123.478 | <0.001 | 37.1 | 99.9 |
| FRics + T | −70.254 | 10 | 162.327 | 123.595 | <0.001 | 35.5 | 99.9 |
| FEvef + T | −70.337 | 10 | 162.492 | 123.760 | <0.001 | 33.9 | 99.9 |
| FDivf + T | −70.423 | 10 | 162.665 | 123.933 | <0.001 | 34.4 | 99.9 |
| HAED + T | −70.445 | 10 | 162.707 | 123.975 | <0.001 | 33.9 | 99.9 |
| FEves + T | −70.476 | 10 | 162.770 | 124.038 | <0.001 | 33.9 | 99.9 |
| MPDed + T | −70.476 | 10 | 162.771 | 124.039 | <0.001 | 33.9 | 99.9 |
| FRicf×T | −65.664 | 14 | 162.917 | 124.185 | <0.001 | 41.1 | 99.9 |
| NNDed×T | −65.853 | 14 | 163.296 | 124.564 | <0.001 | 38.8 | 99.9 |
| RaoQs×T | −65.916 | 14 | 163.421 | 124.689 | <0.001 | 37.8 | 99.9 |
| MPD × T | −66.237 | 14 | 164.065 | 125.332 | <0.001 | 36.9 | 99.9 |
| HEED × T | −66.671 | 14 | 164.932 | 126.199 | <0.001 | 36.5 | 99.9 |
| NRI × T | −66.679 | 14 | 164.948 | 126.216 | <0.001 | 36.5 | 99.9 |
| NTI × T | −67.022 | 14 | 165.634 | 126.902 | <0.001 | 37.4 | 99.9 |
| FDivs × T | −67.470 | 14 | 166.530 | 127.798 | <0.001 | 36.9 | 99.9 |
| NTIed × T | −68.012 | 14 | 167.615 | 128.882 | <0.001 | 35.8 | 99.9 |
| FDisf × T | −68.143 | 14 | 167.876 | 129.144 | <0.001 | 36.5 | 99.9 |
| RaoQf × T | −68.432 | 14 | 168.454 | 129.722 | <0.001 | 36.0 | 99.9 |
| H maxf × T | −68.722 | 14 | 169.034 | 130.302 | <0.001 | 34.8 | 99.9 |
| FEvef × T | −69.058 | 14 | 169.706 | 130.974 | <0.001 | 34.6 | 99.9 |
| FEves × T | −69.170 | 14 | 169.930 | 131.198 | <0.001 | 34.9 | 99.9 |
| LCs × T | −69.377 | 14 | 170.344 | 131.612 | <0.001 | 35.0 | 99.9 |
| HAED × T | −69.433 | 14 | 170.455 | 131.723 | <0.001 | 34.4 | 99.9 |
| FDivf × T | −69.783 | 14 | 171.155 | 132.423 | <0.001 | 34.8 | 99.9 |
| 1 | −80.940 | 5 | 172.356 | 133.624 | <0.001 | 0.0 | 91.1 |
Fixed factors are number of species (S), Shannon's evenness (H’), and phylogenetic diversity (PD, sum of branch lengths; IAC, imbalance of abundance at the clade; EAED, equitability of abundance‐weighted entropic measure of the distribution of evolutionary distinctiveness; MPD, mean pairwise distance; MNND, mean nearest‐neighbor distance; MPDed, weighted mean pairwise distance; MNNDed, weighted mean nearest neighbor distance); and community‐level mean of single functional traits using interspecific and intraspecific functional traits indicated by subscript “f” and “s,” respectively (H max , maximum plant height; LC, leaf carbon content; LN, leaf nitrogen content; LP, leaf phosphorus content; SLA, specific leaf area) or multivariate functional trait indices using interspecific and intraspecific functional traits indicated by subscript “f” and “s,” respectively (FDis, functional distribution; FRic, functional richness; FEve, functional evenness; FDiv, functional divergence; RaoQ, quadratic entropy), and experimental treatments (T: T C = control, T W = warming, T N = nitrogen addition, T WN = warming and nitrogen addition, and T G = livestock‐grazing). Hierarchical random factor is elevation (2,700, 3,200, and 3,400 m), treatment, and plot. Values are shown for the estimated number of model parameters (k), maximum log‐likelihood (LL), and the information‐theoretic Akaike's information criterion corrected for small samples (AICc), change in AICc relative to the top‐ranked model (ΔAICc), AICc weight (wAICc, model probability), and the marginal and total variance explained (R m 2, R c 2) as a measure of the model's goodness of fit.
Table A3.
Spearman's ρ correlation matrix for raw input variables
| S | H’ | PD | IAC | HEED | HAED | MPD | MPDed | MNND | MNNDed | FRicf | FEvef | FDivf | FDisf | RaoQf | H maxf | LC f | LN f | LP f | SLAf | FRics | FEves | FDivs | FDiss | RaoQs | Hmaxs | LCs | LNs | LPs | SLAs | pH | P | N | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H’ | 0.65 | ||||||||||||||||||||||||||||||||
| PD | 0.94 | 0.60 | |||||||||||||||||||||||||||||||
| IAC | −0.22 | −0.41 | −0.18 | ||||||||||||||||||||||||||||||
| HEED | 0.16 | 0.31 | 0.09 | −0.42 | |||||||||||||||||||||||||||||
| HAED | 0.27 | 0.35 | 0.20 | −0.23 | 0.96 | ||||||||||||||||||||||||||||
| MPD | −0.03 | −0.18 | 0.15 | 0.13 | −0.20 | −0.17 | |||||||||||||||||||||||||||
| MPDed | 0.41 | 0.72 | 0.44 | −0.37 | 0.20 | 0.23 | 0.18 | ||||||||||||||||||||||||||
| MNND | −0.62 | −0.45 | −0.43 | 0.18 | −0.26 | −0.33 | 0.39 | −0.22 | |||||||||||||||||||||||||
| MNNDed | −0.33 | −0.11 | −0.24 | −0.05 | −0.04 | −0.07 | 0.23 | 0.23 | 0.54 | ||||||||||||||||||||||||
| FRicf | 0.85 | 0.44 | 0.81 | −0.12 | 0.09 | 0.19 | 0.12 | 0.33 | −0.49 | −0.27 | |||||||||||||||||||||||
| FEvef | −0.27 | −0.14 | −0.26 | 0.02 | 0.08 | 0.03 | −0.15 | −0.25 | 0.10 | 0.06 | −0.29 | ||||||||||||||||||||||
| FDivf | 0.26 | 0.02 | 0.27 | 0.12 | 0.00 | 0.08 | 0.16 | 0.06 | −0.15 | −0.11 | 0.37 | 0.05 | |||||||||||||||||||||
| FDisf | 0.27 | 0.20 | 0.21 | 0.07 | 0.04 | 0.11 | −0.08 | 0.00 | −0.25 | −0.16 | 0.31 | 0.22 | 0.64 | ||||||||||||||||||||
| RaoQf | 0.24 | 0.21 | 0.18 | 0.06 | 0.05 | 0.11 | −0.12 | −0.03 | −0.24 | −0.13 | 0.24 | 0.27 | 0.59 | 0.96 | |||||||||||||||||||
| H maxf | −0.44 | −0.31 | −0.40 | 0.45 | −0.16 | −0.13 | −0.01 | −0.22 | 0.33 | 0.17 | −0.34 | 0.28 | 0.07 | 0.06 | 0.05 | ||||||||||||||||||
| LC f | −0.06 | −0.07 | −0.04 | −0.12 | −0.05 | −0.07 | 0.22 | 0.13 | 0.02 | 0.10 | 0.00 | −0.20 | 0.01 | −0.15 | −0.16 | −0.27 | |||||||||||||||||
| LN f | −0.07 | 0.11 | −0.10 | 0.18 | 0.05 | 0.08 | −0.36 | −0.16 | 0.05 | 0.17 | −0.22 | 0.35 | −0.04 | 0.21 | 0.28 | 0.34 | −0.27 | ||||||||||||||||
| LP f | −0.44 | −0.11 | −0.45 | 0.07 | −0.06 | −0.13 | −0.35 | −0.27 | 0.31 | 0.25 | −0.54 | 0.37 | −0.30 | −0.02 | 0.10 | 0.40 | −0.29 | 0.54 | |||||||||||||||
| SLA f | −0.30 | −0.33 | −0.22 | 0.14 | −0.10 | −0.12 | 0.49 | −0.07 | 0.29 | −0.04 | −0.12 | −0.12 | −0.06 | −0.26 | −0.33 | 0.23 | 0.29 | −0.50 | −0.37 | ||||||||||||||
| FRics | 0.86 | 0.46 | 0.81 | −0.17 | 0.08 | 0.18 | 0.12 | 0.35 | −0.48 | −0.27 | 0.96 | −0.28 | 0.35 | 0.31 | 0.25 | −0.38 | 0.00 | −0.25 | −0.52 | −0.15 | |||||||||||||
| FEves | −0.29 | −0.16 | −0.26 | 0.02 | 0.04 | −0.04 | −0.17 | −0.24 | 0.18 | 0.10 | −0.36 | 0.77 | 0.03 | 0.17 | 0.22 | 0.27 | −0.15 | 0.33 | 0.44 | −0.07 | −0.34 | ||||||||||||
| FDivs | 0.25 | 0.09 | 0.25 | 0.08 | −0.02 | 0.06 | 0.07 | 0.11 | −0.18 | 0.00 | 0.32 | 0.13 | 0.80 | 0.62 | 0.59 | 0.12 | 0.00 | 0.06 | −0.10 | −0.19 | 0.30 | 0.10 | |||||||||||
| FDiss | 0.24 | 0.23 | 0.21 | 0.05 | 0.02 | 0.06 | −0.10 | 0.01 | −0.24 | −0.07 | 0.22 | 0.33 | 0.45 | 0.80 | 0.78 | 0.08 | −0.18 | 0.28 | 0.11 | −0.24 | 0.22 | 0.30 | 0.64 | ||||||||||
| RaoQs | 0.20 | 0.23 | 0.17 | 0.06 | 0.02 | 0.07 | −0.13 | −0.03 | −0.20 | −0.05 | 0.15 | 0.37 | 0.39 | 0.73 | 0.77 | 0.10 | −0.20 | 0.34 | 0.22 | −0.29 | 0.15 | 0.35 | 0.57 | 0.96 | |||||||||
| H maxs | −0.18 | −0.11 | −0.14 | 0.70 | −0.19 | −0.03 | 0.03 | −0.11 | 0.15 | 0.08 | −0.09 | 0.11 | 0.07 | 0.08 | 0.06 | 0.68 | −0.19 | 0.28 | 0.22 | 0.08 | −0.15 | 0.11 | 0.12 | 0.13 | 0.16 | ||||||||
| LC s | −0.09 | −0.02 | −0.08 | −0.12 | 0.05 | 0.00 | 0.05 | 0.07 | 0.02 | 0.02 | −0.11 | 0.05 | −0.17 | −0.22 | −0.23 | −0.21 | 0.69 | −0.12 | −0.07 | 0.16 | −0.11 | 0.05 | −0.15 | −0.05 | −0.02 | −0.12 | |||||||
| LN s | −0.08 | 0.12 | −0.11 | 0.16 | 0.08 | 0.10 | −0.36 | −0.15 | 0.05 | 0.14 | −0.24 | 0.39 | −0.09 | 0.16 | 0.23 | 0.31 | −0.25 | 0.97 | 0.55 | −0.48 | −0.27 | 0.36 | 0.01 | 0.29 | 0.37 | 0.27 | 0.06 | ||||||
| LP s | −0.44 | −0.11 | −0.45 | 0.07 | −0.06 | −0.13 | −0.35 | −0.27 | 0.30 | 0.24 | −0.54 | 0.37 | −0.31 | −0.03 | 0.08 | 0.40 | −0.29 | 0.54 | 1.00 | −0.37 | −0.52 | 0.45 | −0.11 | 0.11 | 0.23 | 0.22 | −0.03 | 0.56 | |||||
| SLA s | −0.14 | 0.03 | −0.07 | −0.05 | −0.01 | −0.03 | 0.33 | 0.18 | 0.15 | −0.02 | −0.02 | −0.21 | −0.07 | −0.17 | −0.22 | −0.09 | 0.25 | −0.46 | −0.31 | 0.64 | −0.03 | −0.24 | −0.09 | −0.08 | −0.11 | −0.09 | 0.10 | −0.46 | −0.32 | ||||
| pH | 0.70 | 0.36 | 0.71 | −0.06 | 0.11 | 0.22 | 0.16 | 0.30 | −0.38 | −0.15 | 0.71 | −0.31 | 0.32 | 0.19 | 0.14 | −0.41 | 0.08 | −0.17 | −0.63 | −0.06 | 0.70 | −0.39 | 0.28 | 0.15 | 0.07 | −0.12 | −0.08 | −0.20 | −0.64 | 0.05 | |||
| P | −0.58 | −0.17 | −0.59 | 0.05 | −0.07 | −0.16 | −0.21 | −0.26 | 0.36 | 0.21 | −0.64 | 0.29 | −0.39 | −0.16 | −0.07 | 0.28 | −0.13 | 0.22 | 0.72 | −0.07 | −0.62 | 0.31 | −0.25 | −0.02 | 0.08 | 0.14 | 0.12 | 0.27 | 0.73 | −0.05 | −0.75 | ||
| N | −0.24 | −0.15 | −0.25 | 0.21 | −0.22 | −0.20 | 0.06 | −0.15 | 0.17 | −0.09 | −0.14 | 0.09 | −0.01 | 0.01 | 0.01 | 0.18 | 0.00 | 0.05 | 0.02 | 0.17 | −0.12 | 0.06 | −0.05 | 0.00 | 0.01 | 0.22 | 0.05 | 0.06 | 0.03 | 0.07 | −0.35 | 0.27 | |
| C | −0.23 | −0.20 | −0.24 | 0.17 | −0.18 | −0.18 | 0.13 | −0.10 | 0.17 | −0.05 | −0.10 | 0.06 | −0.08 | −0.06 | −0.09 | 0.17 | −0.02 | 0.03 | 0.01 | 0.14 | −0.09 | 0.05 | −0.08 | −0.08 | −0.08 | 0.18 | 0.03 | 0.04 | 0.01 | 0.00 | −0.31 | 0.15 | 0.74 |
Shown are species richness (S), Shannon's evenness (H’), phylogenetic diversity (PD, sum of branch lengths; IAC, imbalance of abundance at the clade; EAED, equitability of abundance‐weighted entropic measure of the distribution of evolutionary distinctiveness; MPD, mean pairwise distance; MNND, mean nearest neighbor distance; MPDed, weighted mean pairwise distance; MNNDed, weighted mean nearest neighbor distance); multivariate functional trait indices based on interspecific and intraspecific functional traits indicated by subscript “f” and “s,” respectively (FDis, functional distribution; FRic, functional richness; FEve, functional evenness; FDiv, functional divergence; RaoQ, quadratic entropy), community‐level mean of single functional traits also using interspecific and intraspecific functional traits indicated by subscript “f” and “s,” respectively (H max, maximum plant height; LC, leaf carbon content; LN, leaf nitrogen content; LP, leaf phosphorus content; SLA, specific leaf area).
Table A4.
General linear mixed‐effect model (GLMM) results for biomass production as a function of several fixed factors and a hierarchical random factor
| Model | LL | k | AICc | ΔAICc | wAICc | R m 2 | R c 2 |
|---|---|---|---|---|---|---|---|
| S + IAC + LN + C + T | 57.115 | 13 | −85.145 | 0.000 | 0.291 | 91.8 | 99.9 |
| S + IAC + LN + T | 55.664 | 12 | −84.706 | 0.439 | 0.234 | 91.6 | 99.9 |
| S + IAC + LN + N + T | 56.336 | 13 | −83.588 | 1.557 | 0.134 | 91.7 | 99.9 |
| S + IAC + C + T | 54.529 | 12 | −82.437 | 2.708 | 0.075 | 91.4 | 99.9 |
| S + IAC + T | 53.195 | 11 | −82.189 | 2.955 | 0.066 | 91.3 | 99.9 |
| S + IAC + FDis + C + T | 55.010 | 13 | −80.934 | 4.210 | 0.035 | 91.5 | 99.9 |
| S + IAC + N + T | 53.732 | 12 | −80.843 | 4.302 | 0.034 | 91.3 | 99.9 |
| S + IAC + FDis + T | 53.725 | 12 | −80.829 | 4.316 | 0.034 | 91.3 | 99.9 |
| S + IAC + RaoQ + C + T | 54.906 | 13 | −80.726 | 4.418 | 0.032 | 91.5 | 99.9 |
| S + IAC + RaoQ + T | 53.642 | 12 | −80.661 | 4.483 | 0.031 | 91.3 | 99.9 |
| S + IAC + FDis + N + T | 54.336 | 13 | −79.588 | 5.556 | 0.018 | 91.4 | 99.9 |
| S + IAC + RaoQ + N + T | 54.255 | 13 | −79.425 | 5.719 | 0.017 | 91.4 | 99.9 |
| IAC + NND + LN + C + T | 20.838 | 13 | −12.592 | 72.553 | <0.001 | 78.2 | 98.3 |
| IAC + NND + LN + N + T | 19.234 | 13 | −9.383 | 75.762 | <0.001 | 77.9 | 98.2 |
| IAC + NND + LN + T | 17.491 | 12 | −8.360 | 76.785 | <0.001 | 77.1 | 98.2 |
| IAC + NND + RaoQ + C + T | 17.236 | 13 | −5.386 | 79.758 | <0.001 | 78.5 | 98.4 |
| IAC + NND + FDis + C + T | 16.871 | 13 | −4.657 | 80.487 | <0.001 | 78.8 | 98.3 |
| IAC + NND + RaoQ + N + T | 15.996 | 13 | −2.907 | 82.238 | <0.001 | 78.5 | 98.3 |
| IAC + NND + C + T | 14.646 | 12 | −2.670 | 82.475 | <0.001 | 78.3 | 98.1 |
| IAC + NND + FDis + N + T | 15.593 | 13 | −2.101 | 83.043 | <0.001 | 78.7 | 98.2 |
| IAC + NND + RaoQ + T | 14.180 | 12 | −1.737 | 83.407 | <0.001 | 77.6 | 98.1 |
| IAC + NND + FDis + T | 13.832 | 12 | −1.042 | 84.102 | <0.001 | 77.9 | 98.1 |
| IAC + NND + N + T | 13.113 | 12 | 0.396 | 85.541 | <0.001 | 78.0 | 98.1 |
| IAC + NND + T | 11.641 | 11 | 0.918 | 86.062 | <0.001 | 77.2 | 98.0 |
| IAC + LN + pH + T | 6.846 | 12 | 12.929 | 98.074 | <0.001 | 82.3 | 99.9 |
| IAC + RaoQ + pH + T | 6.242 | 12 | 14.139 | 99.283 | <0.001 | 81.8 | 99.9 |
| IAC + FDis + pH + T | 5.845 | 12 | 14.932 | 99.9.077 | <0.001 | 82.1 | 99.9 |
| IAC + LN + C + T | 2.756 | 12 | 21.109 | 106.254 | <0.001 | 69.0 | 99.9 |
| IAC + pH + T | 1.462 | 11 | 21.276 | 106.420 | <0.001 | 80.8 | 99.9 |
| IAC + RaoQ + C + T | 2.402 | 12 | 21.818 | 106.962 | <0.001 | 70.3 | 99.9 |
| IAC + FDis + C + T | 1.676 | 12 | 23.270 | 108.414 | <0.001 | 70.6 | 99.9 |
| IAC + LN + P + C + T | 2.859 | 13 | 23.367 | 108.511 | <0.001 | 67.5 | 99.9 |
| IAC + RaoQ + P + C + T | 2.428 | 13 | 24.229 | 109.373 | <0.001 | 69.5 | 99.9 |
| IAC + RaoQ + N + T | 0.789 | 12 | 25.045 | 110.189 | <0.001 | 70.2 | 99.9 |
| IAC + FDis + P + C + T | 1.745 | 13 | 25.594 | 110.739 | <0.001 | 69.3 | 99.9 |
| IAC + LN + N + T | 0.252 | 12 | 26.118 | 111.262 | <0.001 | 68.7 | 99.1 |
| IAC + FDis + N + T | −0.088 | 12 | 26.797 | 111.942 | <0.001 | 70.6 | 99.9 |
| IAC + RaoQ + P + N + T | 0.794 | 13 | 27.497 | 112.642 | <0.001 | 69.9 | 99.9 |
| IAC + RaoQ + T | −1.812 | 11 | 27.824 | 112.968 | <0.001 | 68.9 | 99.9 |
| IAC + LN + P + N + T | 0.308 | 13 | 28.468 | 113.613 | <0.001 | 67.3 | 98.6 |
| IAC + LN + T | −2.241 | 11 | 28.682 | 113.827 | <0.001 | 67.1 | 98.1 |
| IAC + FDis + P + N + T | −0.058 | 13 | 29.200 | 114.345 | <0.001 | 69.7 | 99.9 |
| IAC + FDis + T | −2.728 | 11 | 29.656 | 114.800 | <0.001 | 69.2 | 99.9 |
| IAC + RaoQ + P + T | −1.670 | 12 | 29.961 | 115.106 | <0.001 | 70.8 | 99.9 |
| IAC + LN + P + T | −2.219 | 12 | 31.059 | 116.204 | <0.001 | 68.0 | 98.3 |
| IAC + C + T | −3.686 | 11 | 31.572 | 116.716 | <0.001 | 68.6 | 99.9 |
| IAC + FDis + P + T | −2.648 | 12 | 31.919 | 117.063 | <0.001 | 70.7 | 99.9 |
| IAC + P + C + T | −3.352 | 12 | 33.327 | 118.471 | <0.001 | 65.7 | 99.9 |
| IAC + N + T | −6.157 | 11 | 36.515 | 121.659 | <0.001 | 68.2 | 99.9 |
| IAC + P + N + T | −5.938 | 12 | 38.497 | 123.642 | <0.001 | 65.7 | 98.6 |
| IAC + T | −8.457 | 10 | 38.732 | 123.877 | <0.001 | 66.7 | 97.9 |
| IAC + P + T | −8.445 | 11 | 41.091 | 126.235 | <0.001 | 66.1 | 97.9 |
| H max + LN + pH + T | −36.620 | 12 | 99.861 | 185.006 | <0.001 | 65.3 | 99.9 |
| H max + pH + T | −38.365 | 11 | 99.9.930 | 186.075 | <0.001 | 64.4 | 99.9 |
| S + H max + T | −38.574 | 11 | 101.347 | 186.492 | <0.001 | 61.4 | 95.9 |
| S + H max + LN + T | −37.695 | 12 | 102.011 | 187.156 | <0.001 | 60.4 | 96.4 |
| S + H max + C + T | −37.828 | 12 | 102.278 | 187.422 | <0.001 | 61.2 | 96.0 |
| S + H max + LN + C + T | −36.857 | 13 | 102.798 | 187.943 | <0.001 | 60.3 | 96.3 |
| S + H max + N + T | −38.205 | 12 | 103.032 | 188.177 | <0.001 | 61.5 | 95.9 |
| S + H max + LN + N + T | −37.276 | 13 | 103.637 | 188.782 | <0.001 | 60.4 | 96.5 |
| NND + H max + LN + C + T | −37.453 | 13 | 103.991 | 189.136 | <0.001 | 56.3 | 96.3 |
| NND + H max + LN + T | −38.868 | 12 | 104.358 | 189.502 | <0.001 | 55.6 | 96.3 |
| NND + H max + C + T | −39.270 | 12 | 105.162 | 190.307 | <0.001 | 56.4 | 96.0 |
| NND + H max + LN + N + T | −38.097 | 13 | 105.278 | 190.422 | <0.001 | 56.2 | 96.4 |
| NND + H max + T | −40.642 | 11 | 105.484 | 190.629 | <0.001 | 55.7 | 95.9 |
| NND + H max + N + T | −39.902 | 12 | 106.425 | 191.570 | <0.001 | 56.2 | 96.0 |
| H max + LN + C + T | −40.844 | 12 | 108.309 | 193.454 | <0.001 | 51.6 | 99.9 |
| H max + LN + T | −42.684 | 11 | 109.567 | 194.712 | <0.001 | 50.7 | 99.9 |
| H max + LN + N + T | −41.609 | 12 | 109.840 | 194.985 | <0.001 | 51.5 | 99.9 |
| H max + C + T | −42.933 | 11 | 110.066 | 195.211 | <0.001 | 51.3 | 97.2 |
| H’ + H max + LN + C + T | −40.551 | 13 | 110.187 | 195.332 | <0.001 | 51.3 | 99.9 |
| H max + LN + P + C + T | −40.686 | 13 | 110.458 | 195.602 | <0.001 | 53.8 | 99.9 |
| H max + LN + P + T | −42.179 | 12 | 110.980 | 196.125 | <0.001 | 54.6 | 99.9 |
| H max + T | −44.775 | 10 | 111.367 | 196.512 | <0.001 | 50.2 | 98.2 |
| H max + N + T | −43.679 | 11 | 111.558 | 196.702 | <0.001 | 51.1 | 99.9 |
| H + H max + LN + T | −42.557 | 12 | 111.736 | 196.881 | <0.001 | 50.4 | 99.9 |
| H’ + H max + LN + N + T | −41.403 | 13 | 111.891 | 197.036 | <0.001 | 51.3 | 99.9 |
| H max + LN + P + N + T | −41.410 | 13 | 111.905 | 197.050 | <0.001 | 54.0 | 99.9 |
| H max + P + C + T | −42.856 | 12 | 112.334 | 197.478 | <0.001 | 52.9 | 96.9 |
| H’ + H max + C + T | −42.865 | 12 | 112.352 | 197.497 | <0.001 | 51.1 | 97.5 |
| H’ + H max + LN + P + C + T | −40.477 | 14 | 112.544 | 197.688 | <0.001 | 52.8 | 99.9 |
| H max + P + T | −44.432 | 11 | 113.065 | 198.209 | <0.001 | 53.6 | 97.1 |
| H’ + H max + LN + P + T | −42.136 | 13 | 113.357 | 198.501 | <0.001 | 54.1 | 99.9 |
| H’ + H max + T | −44.766 | 11 | 113.733 | 198.877 | <0.001 | 50.1 | 98.3 |
| H max + P + N + T | −43.586 | 12 | 113.793 | 198.938 | <0.001 | 52.8 | 98.4 |
| H’ + H max + N + T | −43.643 | 12 | 113.907 | 199.052 | <0.001 | 50.9 | 99.9 |
| H’ + H max + LN + P + N + T | −41.283 | 14 | 114.157 | 199.301 | <0.001 | 53.2 | 99.9 |
| H’ + H max + P + C + T | −42.821 | 13 | 114.727 | 199.871 | <0.001 | 52.4 | 97.2 |
| H’ + H max + P + T | −44.429 | 12 | 115.480 | 200.624 | <0.001 | 53.8 | 97.1 |
| H’ + H max + P + N + T | −43.573 | 13 | 116.231 | 201.375 | <0.001 | 52.5 | 99.9 |
| LN + pH + T | −57.526 | 11 | 139.252 | 224.396 | <0.001 | 52.2 | 99.9 |
| NND + LN + T | −58.903 | 11 | 142.006 | 227.150 | <0.001 | 44.3 | 99.9 |
| S + LN + T | −58.913 | 11 | 142.027 | 227.171 | <0.001 | 48.2 | 99.9 |
| NND + LN + C + T | −57.908 | 12 | 142.439 | 227.583 | <0.001 | 44.9 | 97.7 |
| S + LN + C + T | −58.424 | 12 | 143.471 | 228.615 | <0.001 | 47.9 | 99.9 |
| NND + LN + N + T | −58.753 | 12 | 144.128 | 229.273 | <0.001 | 44.5 | 99.9 |
| S + LN + N + T | −58.896 | 12 | 144.414 | 229.559 | <0.001 | 48.2 | 99.9 |
| LN + C + T | −62.205 | 11 | 148.611 | 233.755 | <0.001 | 39.4 | 99.9 |
| LN + T | −63.666 | 10 | 149.151 | 234.296 | <0.001 | 38.3 | 99.9 |
| RaoQ + pH + T | −62.497 | 11 | 149.193 | 234.338 | <0.001 | 48.4 | 99.9 |
| S + T | −63.748 | 10 | 149.315 | 234.459 | <0.001 | 46.8 | 99.9 |
| FDis + pH + T | −62.610 | 11 | 149.420 | 234.565 | <0.001 | 48.5 | 99.9 |
| H’ + LN + P + T | −61.491 | 12 | 149.605 | 234.749 | <0.001 | 50.1 | 99.9 |
| S + FDis + T | −62.862 | 11 | 149.923 | 235.068 | <0.001 | 46.9 | 99.9 |
| LN + P + T | −62.889 | 11 | 149.979 | 235.123 | <0.001 | 49.1 | 99.9 |
| S + RaoQ + T | −62.967 | 11 | 150.134 | 235.278 | <0.001 | 46.6 | 99.9 |
| H’ + LN + C + T | −61.896 | 12 | 150.414 | 235.558 | <0.001 | 40.0 | 99.9 |
| H’ + LN + T | −63.172 | 11 | 150.544 | 235.689 | <0.001 | 39.1 | 99.9 |
| LN + P + C + T | −62.024 | 12 | 150.671 | 235.815 | <0.001 | 41.8 | 99.9 |
| pH + T | −64.576 | 10 | 150.970 | 236.114 | <0.001 | 47.1 | 99.9 |
| LN + N + T | −63.394 | 11 | 150.988 | 236.133 | <0.001 | 38.7 | 99.9 |
| S + C + T | −63.454 | 11 | 151.108 | 236.253 | <0.001 | 46.6 | 99.9 |
| H’ + LN + P + C + T | −61.099 | 13 | 151.283 | 236.427 | <0.001 | 50.4 | 99.9 |
| S + N + T | −63.748 | 11 | 151.696 | 236.84 | <0.001 | 46.8 | 99.9 |
| S + FDis + C + T | −62.554 | 12 | 151.730 | 236.875 | <0.001 | 46.7 | 99.9 |
| NND + RaoQ + T | −63.844 | 11 | 151.888 | 237.033 | <0.001 | 41.7 | 99.9 |
| S + RaoQ + C + T | −62.660 | 12 | 151.942 | 237.087 | <0.001 | 46.3 | 99.9 |
| NND + FDis + T | −63.922 | 11 | 152.045 | 237.189 | <0.001 | 42.0 | 99.9 |
| H’ + LN + P + N + T | −61.490 | 13 | 152.064 | 237.209 | <0.001 | 50.1 | 99.9 |
| S + FDis + N + T | −62.854 | 12 | 152.330 | 237.475 | <0.001 | 46.9 | 99.9 |
| NND + T | −65.273 | 10 | 152.364 | 237.509 | <0.001 | 41.1 | 99.9 |
| LN + P + N + T | −62.874 | 12 | 152.369 | 237.513 | <0.001 | 49.1 | 99.9 |
| H’ + LN + N + T | −62.944 | 12 | 152.510 | 237.654 | <0.001 | 39.4 | 99.9 |
| S + RaoQ + N + T | −62.960 | 12 | 152.542 | 237.687 | <0.001 | 46.6 | 99.9 |
| NND + RaoQ + C + T | −63.067 | 12 | 152.757 | 237.901 | <0.001 | 42.2 | 99.9 |
| NND + FDis + C + T | −63.112 | 12 | 152.845 | 237.99 | <0.001 | 42.5 | 99.9 |
| NND + C + T | −64.367 | 11 | 152.934 | 238.078 | <0.001 | 41.6 | 99.9 |
| NND + RaoQ + N + T | −63.714 | 12 | 154.050 | 239.194 | <0.001 | 41.9 | 99.9 |
| NND + FDis + N + T | −63.784 | 12 | 154.190 | 239.334 | <0.001 | 42.2 | 99.9 |
| NND + N + T | −65.159 | 11 | 154.519 | 239.663 | <0.001 | 41.3 | 99.9 |
| H’ + RaoQ + P + T | −65.054 | 12 | 156.730 | 241.874 | <0.001 | 47.4 | 99.9 |
| RaoQ + P + T | −66.341 | 11 | 156.882 | 242.027 | <0.001 | 46.3 | 99.9 |
| H’ + FDis + P + T | −65.162 | 12 | 156.947 | 242.091 | <0.001 | 47.3 | 99.9 |
| RaoQ + T | −67.614 | 10 | 157.047 | 242.192 | <0.001 | 36.3 | 99.9 |
| RaoQ + C + T | −66.522 | 11 | 157.245 | 242.389 | <0.001 | 37.2 | 99.9 |
| FDis + P + T | −66.568 | 11 | 157.336 | 242.481 | <0.001 | 46.1 | 99.9 |
| FDis + T | −67.792 | 10 | 157.403 | 242.547 | <0.001 | 36.5 | 99.9 |
| FDis + C + T | −66.634 | 11 | 157.469 | 242.613 | <0.001 | 37.4 | 99.9 |
| H’ + RaoQ + T | −66.890 | 11 | 157.980 | 243.125 | <0.001 | 37.4 | 99.9 |
| H’ + FDis + T | −66.955 | 11 | 158.109 | 243.254 | <0.001 | 37.7 | 99.9 |
| H’ + RaoQ + C + T | −65.963 | 12 | 158.549 | 243.693 | <0.001 | 38.1 | 99.9 |
| H’ + FDis + C + T | −65.991 | 12 | 158.603 | 243.747 | <0.001 | 38.4 | 99.9 |
| RaoQ + P + C + T | −66.035 | 12 | 158.693 | 243.837 | <0.001 | 46.6 | 99.9 |
| H’ + RaoQ + P + C + T | −64.863 | 13 | 158.812 | 243.956 | <0.001 | 47.5 | 99.9 |
| H’ + P + T | −67.316 | 11 | 158.831 | 243.976 | <0.001 | 45.5 | 99.9 |
| RaoQ + N + T | −67.379 | 11 | 158.958 | 244.103 | <0.001 | 36.6 | 99.9 |
| H’ + FDis + P + C + T | −64.937 | 13 | 158.959 | 244.104 | <0.001 | 47.5 | 99.9 |
| FDis + P + C + T | −66.201 | 12 | 159.024 | 244.169 | <0.001 | 46.4 | 99.9 |
| H’ + RaoQ + P + N + T | −65.053 | 13 | 159.191 | 244.336 | <0.001 | 47.4 | 99.9 |
| FDis + N + T | −67.540 | 11 | 159.280 | 244.425 | <0.001 | 36.8 | 99.9 |
| RaoQ + P + N + T | −66.331 | 12 | 159.283 | 244.427 | <0.001 | 46.3 | 99.9 |
| H’ + FDis + P + N + T | −65.159 | 13 | 159.404 | 244.548 | <0.001 | 47.3 | 99.9 |
| H’ + T | −68.852 | 10 | 159.523 | 244.667 | <0.001 | 36.1 | 99.9 |
| FDis + P + N + T | −66.548 | 12 | 159.719 | 244.863 | <0.001 | 46.2 | 99.9 |
| H’ + C + T | −67.783 | 11 | 159.765 | 244.91 | <0.001 | 37.0 | 99.9 |
| H’ + RaoQ + N + T | −66.703 | 12 | 160.028 | 245.172 | <0.001 | 37.7 | 99.9 |
| C + T | −69.120 | 10 | 160.058 | 245.203 | <0.001 | 35.1 | 99.9 |
| H’ + FDis + N + T | −66.758 | 12 | 160.137 | 245.281 | <0.001 | 38.0 | 99.9 |
| H’ + P + C + T | −67.013 | 12 | 160.648 | 245.792 | <0.001 | 45.8 | 99.9 |
| H’ + P + N + T | −67.315 | 12 | 161.252 | 246.396 | <0.001 | 45.5 | 99.9 |
| H’ + N + T | −68.691 | 11 | 161.582 | 246.727 | <0.001 | 36.4 | 99.9 |
| P + T | −69.946 | 10 | 161.710 | 246.855 | <0.001 | 43.3 | 99.9 |
| P + C + T | −69.089 | 11 | 162.377 | 247.522 | <0.001 | 36.2 | 99.9 |
| N + T | −70.296 | 10 | 162.410 | 247.555 | <0.001 | 34.3 | 99.9 |
| P + N + T | −69.926 | 11 | 164.052 | 249.197 | <0.001 | 43.3 | 99.9 |
| 1 | −80.940 | 5 | 172.356 | 257.501 | <0.001 | 0.0 | 91.1 |
Fixed factors are number of species (S), Shannon's evenness (H’), and phylogenetic diversity (IAC, imbalance of abundance at the clade; MNND, mean nearest neighbor distance), and community‐level mean of single functional traits (H max, plot‐specific maximum plant height; LN, mean leaf nitrogen content value for individual species used for all plots where the species is found) or multivariate functional trait indices (RaoQ, Quadratic entropy; FDis, Functional dispersion: weighted distances from a weighted centroid in multitrait space), and experimental treatments (T: T C = control, T W = warming, T N = nitrogen addition, T WN = warming and nitrogen addition, and T G = livestock‐grazing), and soil resources (C, soil carbon content; N, soil total nitrogen content; P, soil total phosphorus content). Hierarchical random factor is elevation (2700 m, 3200 m and 3400 m), treatment, and plot. Values are shown for the estimated number of model parameters (k), maximum log‐likelihood (LL), and the information‐theoretic Akaike's information criterion corrected for small samples (AICc), change in AICc relative to the top‐ranked model (ΔAICc), AICc weight (wAICc, model probability), and the marginal and total variance explained (Rm 2, Rc 2) as a measure of the model's goodness of fit.
Liu J, Liu D, Xu K, et al. Biodiversity explains maximum variation in productivity under experimental warming, nitrogen addition, and grazing in mountain grasslands. Ecol Evol. 2018;8:10094–10112. 10.1002/ece3.4483
REFERENCES
- Adler, P. B. , Seabloom, E. W. , Borer, E. T. , Hillebrand, H. , Hautier, Y. , Hector, A. , … Yang, L. H. (2011). Productivity is a poor predictor of plant species richness. Science, 333(6050), 1750–1753. 10.1126/science.1204498 [DOI] [PubMed] [Google Scholar]
- Albert, C. H. , de Bello, F. , Boulangeat, I. , Pellet, G. , Lavorel, S. , & Thuiller, W. (2012). On the importance of intraspecific variability for the quantification of functional diversity. Oikos, 121(1), 116–126. 10.1111/j.1600-0706.2011.19672.x [DOI] [Google Scholar]
- Ali, A. , & Mattsson, E. (2017). Disentangling the effects of species diversity, and intraspecific and interspecific tree size variation on aboveground biomass in dry zone homegarden agroforestry systems. Science of the Total Environment, 598, 38–48. 10.1016/j.scitotenv.2017.04.131 [DOI] [PubMed] [Google Scholar]
- Aspinwall, M. J. , Loik, M. E. , Resco de Dios, V. , Tjoelker, M. G. , Payton, P. R. , & Tissue, D. T. (2015). Utilizing intraspecific variation in phenotypic plasticity to bolster agricultural and forest productivity under climate change. Plant Cell and Environment, 38(9), 1752–1764. 10.1111/pce.12424 [DOI] [PubMed] [Google Scholar]
- Balvanera, P. , Pfisterer, A. B. , Buchmann, N. , He, J.‐S. , Nakashizuka, T. , Raffaelli, D. , & Schmid, B. (2006). Quantifying the evidence for biodiversity effects on ecosystem functioning and services. Ecology Letters, 9(10), 1146–1156. 10.1111/j.1461-0248.2006.00963.x [DOI] [PubMed] [Google Scholar]
- Borer, E. T. , Seabloom, E. W. , Gruner, D. S. , Harpole, W. S. , Hillebrand, H. , Lind, E. M. , … Yang, L. H. (2014). Herbivores and nutrients control grassland plant diversity via light limitation. Nature, 508(7497), 517–520. 10.1038/nature13144 [DOI] [PubMed] [Google Scholar]
- Botta‐Dukat, Z. (2005). Rao's quadratic entropy as a measure of functional diversity based on multiple traits. Journal of Vegetation Science, 16(5), 533–540. 10.1111/j.1654-1103.2005.tb02393.x [DOI] [Google Scholar]
- Burnham, K. P. , & Anderson, D. R. (2002). Model selection and multimodel inference: A practical information‐theoretic approach, 2nd ed New York, NY: Springer‐Verlag. [Google Scholar]
- Burnham, K. P. , & Anderson, D. R. (2004). Understanding AIC and BIC in model selection. Sociological Methods and Research, 33, 261–304. 10.1177/0049124104268644 [DOI] [Google Scholar]
- Cadotte, M. W. (2013). Experimental evidence that evolutionarily diverse assemblages result in higher productivity. Proceedings of the National Academy of Sciences of the United States of America, 110(22), 8996–9000. 10.1073/pnas.1301685110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadotte, M. W. (2017). Functional traits explain ecosystem function through opposing mechanisms. Ecology Letters, 20(8), 989–996. 10.1111/ele.12796 [DOI] [PubMed] [Google Scholar]
- Cadotte, M. W. , Cavender‐Bares, J. , Tilman, D. , & Oakley, T. H. (2009). Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLoS ONE, 4(5), e5695 10.1371/journal.pone.0005695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadotte, M. W. , Davies, T. J. , & Peres‐Neto, P. R. (2017). Why phylogenies do not always predict ecological differences. Ecological Monographs, 87(4), 535–551. 10.1002/ecm.1267 [DOI] [Google Scholar]
- Cadotte, M. W. , Davies, T. J. , Regetz, J. , Kembel, S. W. , Cleland, E. , & Oakley, T. H. (2010). Phylogenetic diversity metrics for ecological communities: Integrating species richness, abundance and evolutionary history. Ecology Letters, 13(1), 96–105. 10.1111/j.1461-0248.2009.01405.x [DOI] [PubMed] [Google Scholar]
- Cardinale, B. J. , Srivastava, D. S. , Duffy, J. E. , Wright, J. P. , Downing, A. L. , Sankaran, M. , & Jouseau, C. (2006). Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature, 443(7114), 989–992. 10.1038/nature05202 [DOI] [PubMed] [Google Scholar]
- Chapin, F. S. , Zavaleta, E. S. , Eviner, V. T. , Naylor, R. L. , Vitousek, P. M. , Reynolds, H. L. , … Díaz, S. (2000). Consequences of changing biodiversity. Nature, 405(6783), 234–242. 10.1038/35012241 [DOI] [PubMed] [Google Scholar]
- Chesson, P. L. , & Warner, R. R. (1981). Environmental variability promotes coexistence in lottery competitive systems. American Naturalist, 117(6), 923–943. 10.1086/283778 [DOI] [Google Scholar]
- Cingolani, A. M. , Cabido, M. , Gurvich, D. E. , Renison, D. , & Diaz, S. (2007). Filtering processes in the assembly of plant communities: Are species presence and abundance driven by the same traits? Journal of Vegetation Science, 18(6), 911–920. 10.1111/j.1654-1103.2007.tb02607.x [DOI] [Google Scholar]
- Cingolani, A. M. , Noy‐Meir, I. , & Diaz, S. (2005). Grazing effects on rangeland diversity: A synthesis of contemporary models. Ecological Applications, 15(2), 757–773. 10.1890/03-5272 [DOI] [Google Scholar]
- Cornwell, W. K. , Schwilk, D. W. , & Ackerly, D. D. (2006). A trait‐based test for habitat filtering: Convex hull volume. Ecology, 87, 1465–1471. 10.1890/0012-9658(2006)87[1465:ATTFHF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biological Conservation, 61, 1–10. 10.1016/0006-3207(92)91201-3 [DOI] [Google Scholar]
- Flynn, D. F. B. , Mirotchnick, N. , Jain, M. , Palmer, M. I. , & Naeem, S. (2011). Functional and phylogenetic diversity as predictors of biodiversity‐ecosystem‐function relationships. Ecology, 92(8), 1573–1581. 10.1890/10-1245.1 [DOI] [PubMed] [Google Scholar]
- Fridley, J. D. (2002). Resource availability dominates and alters the relationship between species diversity and ecosystem productivity in experimental plant communities. Oecologia, 132(2), 271–277. 10.1007/s00442-002-0965-x [DOI] [PubMed] [Google Scholar]
- Fridley, J. D. (2003). Diversity effects on production in different light and fertility environments: An experiment with communities of annual plants. Journal of Ecology, 91(3), 396–406. 10.1046/j.1365-2745.2003.00775.x [DOI] [Google Scholar]
- Gough, L. , Grace, J. B. , & Taylor, K. L. (1994). The relationship between species richness and community biomass: The importance of environmental variables. Oikos, 70(2), 271–279. 10.2307/3545638 [DOI] [Google Scholar]
- Guindon, S. , Dufayard, J.‐F. , Lefort, V. , Anisimova, M. , Hordijk, W. , & Gascuel, O. (2010). New algorithms and methods to estimate maximum‐likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology, 59(3), 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hautier, Y. , Niklaus, P. A. , & Hector, A. (2009). Competition for light causes plant biodiversity loss after eutrophication. Science, 324(5927), 636–638. 10.1126/science.1169640 [DOI] [PubMed] [Google Scholar]
- Hector, A. , Joshi, J. , Scherer‐Lorenzen, M. , Schmid, B. , Spehn, E. M. , Wacker, L. , … Troumbis, A. Y. (2007). Biodiversity and ecosystem functioning: Reconciling the results of experimental and observational studies. Functional Ecology, 21(5), 998–1002. 10.1111/j.1365-2435.2007.01308.x [DOI] [Google Scholar]
- Hector, A. , Schmid, B. , Beierkuhnlein, C. , Caldeira, M. C. , Diemer, M. , Dimitrakopoulos, P. G. , … Read, D. J. (1999). Plant diversity and productivity experiments in European grasslands. Science, 286(5442), 1123–1127. 10.1126/science.286.5442.1123 [DOI] [PubMed] [Google Scholar]
- Hodapp, D. , Hillebrand, H. , Blasius, B. , & Ryabov, A. B. (2016). Environmental and trait variability constrain community structure and the biodiversity‐productivity relationship. Ecology, 97(6), 1463–1474. 10.1890/15-0730.1 [DOI] [PubMed] [Google Scholar]
- Hooper, D. U. , Chapin, F. S. , Ewel, J. J. , Hector, A. , Inchausti, P. , Lavorel, S. , … Wardle, D. A. (2005). Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecological Monographs, 75(1), 3–35. 10.1890/04-0922 [DOI] [Google Scholar]
- Horn, H. S. , & Macarthur, R. H. (1972). Competition among fugitive species in a harlequin environment. Ecology, 53(4), 749–752. 10.2307/1934797 [DOI] [Google Scholar]
- Jiang, L. , Wan, S. , & Li, L. (2009). Species diversity and productivity: Why do results of diversity‐manipulation experiments differ from natural patterns? Journal of Ecology, 97(4), 603–608. 10.1111/j.1365-2745.2009.01503.x [DOI] [Google Scholar]
- Jung, V. , Albert, C. H. , Violle, C. , Kunstler, G. , Loucougaray, G. , & Spiegelberger, T. (2014). Intraspecific trait variability mediates the response of subalpine grassland communities to extreme drought events. Journal of Ecology, 102(1), 45–53. 10.1111/1365-2745.12177 [DOI] [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorel, S. , & Garnier, E. (2002). Predicting changes in community composition and ecosystem functioning from plant traits: Revisiting the Holy Grail. Functional Ecology, 16(5), 545–556. 10.1046/j.1365-2435.2002.00664.x [DOI] [Google Scholar]
- Leps, J. , de Bello, F. , Smilauer, P. , & Dolezal, J. (2011). Community trait response to environment: Disentangling species turnover vs intraspecific trait variability effects. Ecography, 34(5), 856–863. 10.1111/j.1600-0587.2010.06904.x [DOI] [Google Scholar]
- Levins, R. , & Culver, D. (1971). Regional coexistence of species and competition between rare species. Proceedings of the National Academy of Sciences of the United States of America, 68(6), 1246–1248. 10.1073/pnas.68.6.1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Yan, H.‐F. , Newmaster, S. G. , Pei, N. , Ragupathy, S. , & Ge, X.‐J. (2015). The use of DNA barcoding as a tool for the conservation biogeography of subtropical forests in China. Diversity and Distributions, 21(2), 188–199. 10.1111/ddi.12276 [DOI] [Google Scholar]
- Liu, J. , Zhang, X. , Song, F. , Zhou, S. , Cadotte, M. W. , & Bradshaw, C. J. A. (2015). Explaining maximum variation in productivity requires phylogenetic diversity and single functional traits. Ecology, 96(1), 176–183. 10.1890/14-1034.1 [DOI] [PubMed] [Google Scholar]
- Luo, Y.‐H. , Liu, J. , Tan, S.‐L. , Cadotte, M. W. , Wang, Y.‐H. , Xu, K. , … Gao, L. M. (2016). Trait‐based community assembly along an elevational gradient in subalpine forests: Quantifying the roles of environmental factors in inter‐ and intraspecific variability. PLoS ONE, 11(5), e0155749 10.1371/journal.pone.0155749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion, G. M. , Henry, G. H. R. , Freckman, D. W. , Johnstone, J. , Jones, G. , Jones, M. H. , … Virginia, R. A. (1997). Open‐top designs for manipulating field temperature in high‐latitude ecosystems. Global Change Biology, 3, 20–32. 10.1111/j.1365-2486.1997.gcb136.x [DOI] [Google Scholar]
- Nakagawa, S. , & Schielzeth, H. (2013). A general and simple method for obtaining R 2 from generalized linear mixed‐effects models. Methods in Ecology and Evolution, 4(2), 133–142. 10.1111/j.2041-210x.2012.00261.x [DOI] [Google Scholar]
- Nicotra, A. B. , Atkin, O. K. , Bonser, S. P. , Davidson, A. M. , Finnegan, E. J. , Mathesius, U. , … van Kleunen, M. (2010). Plant phenotypic plasticity in a changing climate. Trends in Plant Science, 15(12), 684–692. 10.1016/j.tplants.2010.09.008 [DOI] [PubMed] [Google Scholar]
- Paradis, E. , Claude, J. , & Strimmer, K. (2004). APE: Analyses of phylogenetics and evolution in R language. Bioinformatics, 20, 289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Partel, M. , Laanisto, L. , & Zobel, M. (2007). Contrasting plant productivity‐diversity relationships across latitude: The role of evolutionary history. Ecology, 88(5), 1091–1097. 10.1890/06-0997 [DOI] [PubMed] [Google Scholar]
- R Core Team (2016). R: A language and environment for statistical computing. Vienna, Austria: R Foundation of Statistical Computing. [Google Scholar]
- Roscher, C. , Schumacher, J. , Gubsch, M. , Lipowsky, A. , Weigelt, A. , Buchmann, N. , … Schulze, E. D. (2012). Using plant functional traits to explain diversity‐productivity relationships. PLoS ONE, 7(5), e5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, T. , Kohli, L. , Rihm, B. , & Achermann, B. (2013). Nitrogen deposition is negatively related to species richness and species composition of vascular plants and bryophytes in Swiss mountain grassland. Agriculture Ecosystems & Environment, 178, 121–126. 10.1016/j.agee.2013.07.002 [DOI] [Google Scholar]
- Schliep, K. P. (2011). phangorn: Phylogenetic analysis in R. Bioinformatics, 27, 592–593. 10.1093/bioinformatics/btq706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabloom, E. W. , Kinkel, L. , Borer, E. T. , Hautier, Y. , Montgomery, R. A. , & Tilman, D. (2017). Food webs obscure the strength of plant diversity effects on primary productivity. Ecology Letters., 20(4), 505–512. 10.1111/ele.12754 [DOI] [PubMed] [Google Scholar]
- Sparks, D. L. , Page, A. L. , Loeppert, P. A. , Soltanpour, P. N. , Tabatabai, M. A. , Johnston, C. T. , et al. (1996). Methods of soil analysis Part 3: Chemical methods. Madison, WI: Soil Science Society of America and American Society of Agronomy. [Google Scholar]
- Steudel, B. , Hector, A. , Friedl, T. , Loefke, C. , Lorenz, M. , Wesche, M. , … Gessner, M. (2012). Biodiversity effects on ecosystem functioning change along environmental stress gradients. Ecology Letters, 15(12), 1397–1405. 10.1111/j.1461-0248.2012.01863.x [DOI] [PubMed] [Google Scholar]
- Tilman, D. , Isbell, F. , & Cowles, J. M. (2014). Biodiversity and ecosystem functioning in Futuyma DJ, Editor. Annual Review of Ecology, Evolution, and Systematics, 45, 471–493. 10.1146/annurev-ecolsys-120213-091917 [DOI] [Google Scholar]
- Tilman, D. , Reich, P. B. , & Isbell, F. (2012). Biodiversity impacts ecosystem productivity as much as resources, disturbance, or herbivory. Proceedings of the National Academy of Sciences of the United States of America, 109(26), 10394–10397. 10.1073/pnas.1208240109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman, D. , Reich, P. B. , Knops, J. , Wedin, D. , Mielke, T. , & Lehman, C. (2001). Diversity and productivity in a long‐term grassland experiment. Science, 294(5543), 843–845. 10.1126/science.1060391 [DOI] [PubMed] [Google Scholar]
- Tilman, D. , Wedin, D. , & Knops, J. (1996). Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nature, 379, 718–720. 10.1038/379718a0 [DOI] [Google Scholar]
- Urban, M. C. (2015). Accelerating extinction risk from climate change. Science, 348(6234), 571–573. 10.1126/science.aaa4984 [DOI] [PubMed] [Google Scholar]
- Valladares, F. , Gianoli, E. , & Gomez, J. M. (2007). Ecological limits to plant phenotypic plasticity. New Phytologist, 176(4), 749–763. 10.1111/j.1469-8137.2007.02275.x [DOI] [PubMed] [Google Scholar]
- Villeger, S. , Mason, N. W. H. , & Mouillot, D. (2008). New multidimensional functional diversity indices for a multifaceted framework in functional ecology. Ecology, 89(8), 2290–2301. 10.1890/07-1206.1 [DOI] [PubMed] [Google Scholar]
- Webb, C. O. , Ackerly, D. D. , McPeek, M. A. , & Donoghue, M. J. (2002). Phylogenies and community ecology. Annual Review of Ecology and Systematics, 33, 475–505. 10.1146/annurev.ecolsys.33.010802.150448 [DOI] [Google Scholar]
- Weiher, E. , van der Werf, A. , Thompson, K. , Roderick, M. , Garnier, E. , & Eriksson, O. (1999). Challenging Theophrastus: A common core list of plant traits for functional ecology. Journal of Vegetation Science, 10(5), 609–620. 10.2307/3237076 [DOI] [Google Scholar]
- Wright, I. J. , Reich, P. B. , Westoby, M. , Ackerly, D. D. , Baruch, Z. , Bongers, F. , … Villar, R. (2004). The worldwide leaf economics spectrum. Nature, 428, 821–827. 10.1038/nature02403 [DOI] [PubMed] [Google Scholar]
- Zavaleta, E. S. , & Hulvey, K. B. (2007). Realistic variation in species composition affects grassland production, resource use and invasion resistance. Plant Ecology, 188(1), 39–51. [Google Scholar]
- Zhou, X. , Guo, Z. , Zhang, P. , Li, H. , Chu, C. , Li, X. , & Du, G. (2017). Different categories of biodiversity explain productivity variation after fertilization in a Tibetan alpine meadow community. Ecology and Evolution, 7(10), 3464–3474. 10.1002/ece3.2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.5b02c11


