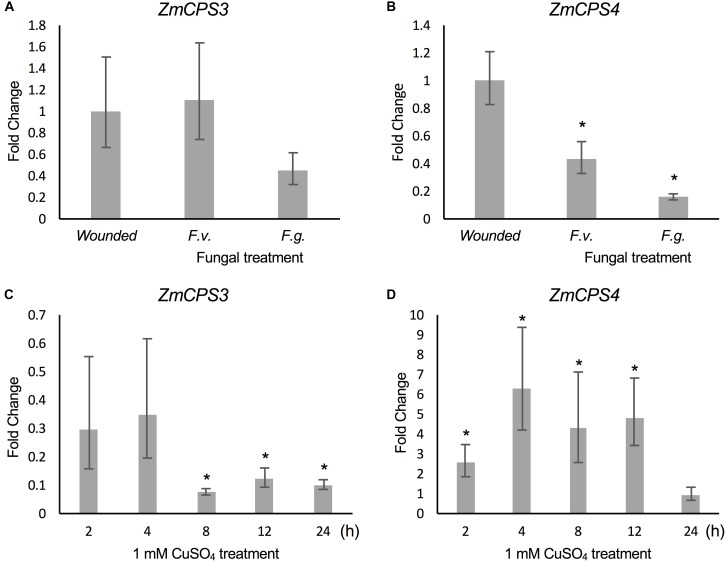
FIGURE 6.
Transcript abundance of ZmCPS3 and ZmCPS4 under biotic and abiotic stress. Transcript abundance was measured by qPCR and is depicted as average fold change (2-ΔΔCt) of the transcript abundance of ZmCPS3 (A) and ZmCPS4 (B) in Mo17 roots incision-inoculated with live conidia, separately with Fusarium verticillioides (F.v.) and F. graminearum (F.g.). Plants treated by incision and application of water only were used as controls. Treatments occurred in 53-days-old plants with tissue harvests 7 days later (n = 4). Average fold change (2-ΔΔCt) of the transcript abundance of ZmCPS3 (C) and ZmCPS4 (D) in Golden Queen roots treated with 1 mM CuSO4 and sampled over 24 h as compared to a water-treated control (n = 3). A fold change of one is equal to no change from the control. Transcript abundance was normalized to the maize internal reference gene EF1α. Error bars represent propagated standard error of the mean fold change of the biological replicates. Asterisks indicate significant change compared to control with P-value < 0.05 as measured with two-tailed Student’s t-tests. A Dunnett’s test was used for fungal treated samples to compare to the single wounded control.