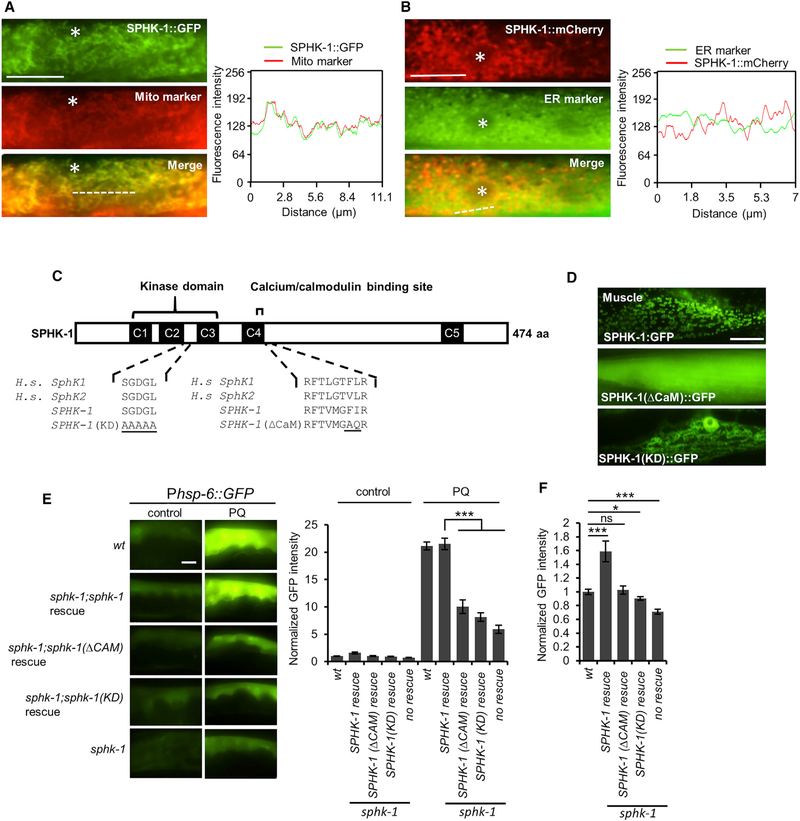
Figure 3. Mitochondrial Localization and Kinase Activity of SPHK-1 Are Essential for sphk-1-Mediated UPRmt Activation.
(A) Co-localization of SPHK-1::GFP fusion proteins with the outer mitochondrial membrane marker TOMM-20::mCherry in the young adult intestines of sphk-1 mutants. Asterisk indicates an intestinal nucleus (left). Cross-sectional fluorescence intensity graph of GFP and mCherry corresponding to the dotted line at left (right).
(B) Co-localization of intestinal SPHK-1::mCherry fusion proteins with the ER marker GFP::C34B2.10. Asterisk indicates intestinal nucleus (left). Cross-sectionalfluorescence intensity graph of GFP and mCherry corresponding to the dotted line at left (right).
(C) Predicted structure of SPHK-1 showing the kinase domain and calmodulin binding motif (CaM). C1–C5 denotes conserved domains found in human (H.s.) SphK1 and SphK2 proteins. Amino acid substitutions made to generate the kinase dead (KD) and ▵CaM SPHK-1 variants are underlined.
(D) Representative images of sphk-1 mutants expressing SPHK-1::GFP, SPHK-1(▵CaM)::GFP, and SPHK-1(KD)::GFP in muscles.
(E) Representative images and quantification of Phsp-6::GFP fluorescence in the indicated strains in the absence or presence of paraquat (for 24 hr). Rescue denotes sphk-1 mutants expressing the indicated sphk-1 cDNA transgenes under control of the intestinal ges-1 promoter.
(F) Average fluorescence of intestinal Phsp-6::GFP expressed in the indicated strains in the absence of stress is quantified.
The sample sizes (n) and ± SEMs are listed in Table S1. Scale bar represents 10 μm. Error bars indicate ± SEMs. Student’s t test; *p < 0.05, ***p < 0.001.