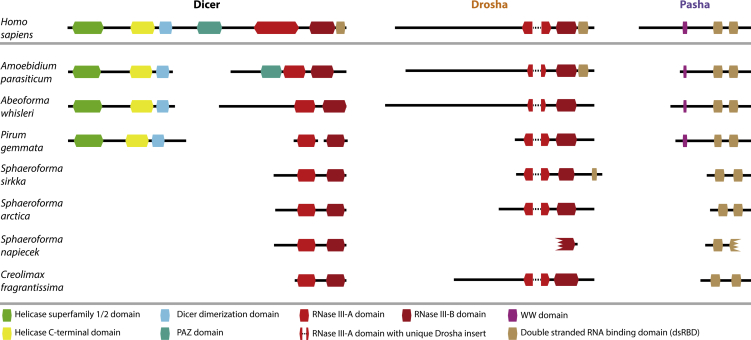
Figure 2.
Comparison between the Domain Composition of the Human and Ichthyosporean miRNA Biogenesis Machinery
The domain composition of the ichthyosporean Dicer, Drosha, and Pasha sequences discovered in the reciprocal BLAST searches was compared against their human counterparts (Dicer [DICER1; Q9UPY3], Drosha [Q9NRR4], and Pasha [DGCR8; Q8WYQ5]), as annotated in InterPro [23]. The sequences identified in the reciprocal BLAST searches were annotated using InterProScan5 [23] and CD-Search [24] and by comparing sequence alignments and secondary structures (see STAR Methods). All domains were identified by both InterProScan and CD-Search annotation programs except the following: Dicer dimerization domain of A. parasiticum; WW domains of A. parasiticum, A. whisleri, and P. gemmata; dsRBD domains of S. sirkka Drosha and of Pasha in P. gemmata, S. arctica, and C. fragrantissima (C-terminal domain only), which were identified by CD-Search only; and the N-terminal dsRBD domains of Pasha in P. gemmata and C. fragrantissima, which were identified by InterProScan only. The RNase III-A domains of S. sirkka, S. arctica, and S. napiecek Dicer were identified using an alignment and structural modeling approach as described in STAR Methods. In addition, the single RNaseIII domain of S. napiecek Drosha was only identified by InterProScan. Incomplete domains are indicated by a jagged border. All boxes and lines are drawn to scale according to their InterProScan annotation (in cases where InterProScan did not identify a domain, the size was chosen based on the homologous domain from a closely related sequence). Except for C. fragrantissima, all genes are from de novo assembled transcriptome data; hence, the many short contigs and aberrant domains are likely due to incomplete assemblies.
See also Figure S1.