Figure 7: Tsg6 −/− and bikunin −/− mice fail to initiate gut rotation.
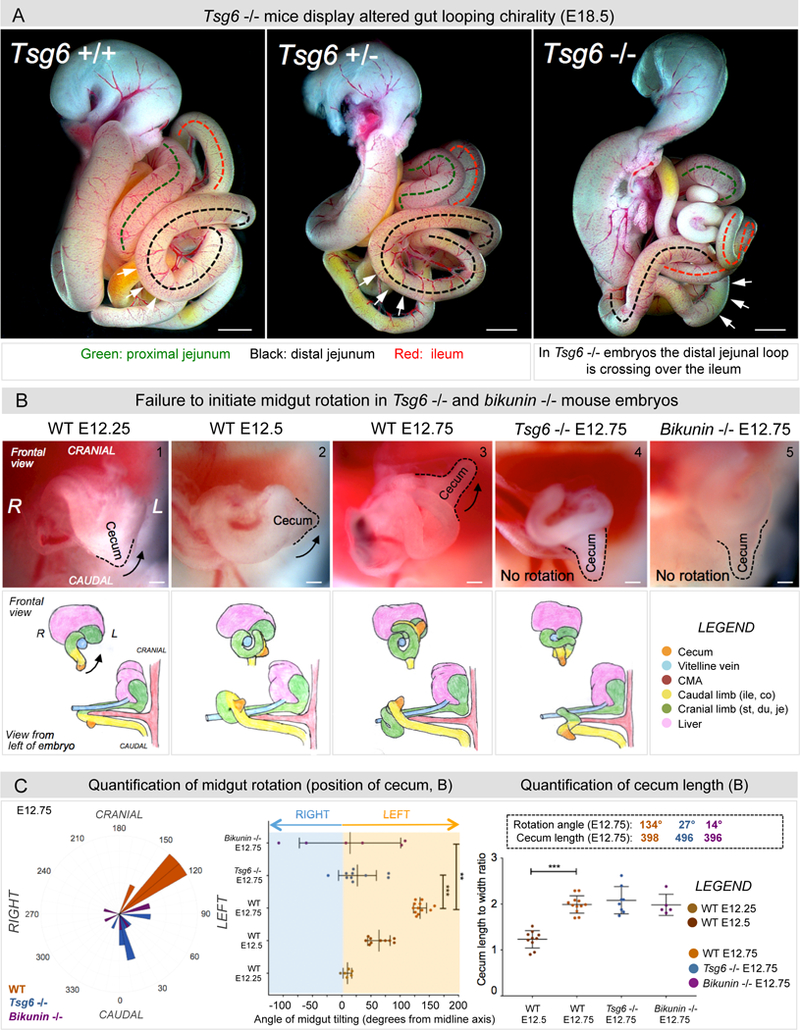
A E18.5 embryos: In Tsg6 −/− embryos the distal jejunal loop crosses over the ileum, indicating malrotation (p = 0.0172 for WT vs Tsg6 −/− embryos, n = 0/7 WT, n = 4/5 Tsg6 −/−). B Midgut rotation in mice initiates at E12 and is restricted to the midgut apex. Unlike WT embryos (panel 1–3), Tsg6 −/− and bikunin −/− embryos fail to initiate rotation as seen by the cecal position (compare panel 3 with 4–5). C Left panel: Polar histogram of cecal angle with respect to the cranial-caudal axis also shown as a dot plot (Middle panel). Right panel: Cecum length-to-width ratios indicating no delay in gut development. Numbers within boxes summarize mean angles of cecum rotation and length at E12.25–12.75 (p = 0.00019 for WT E12.75 vs Tsg6 −/− E12.75, p = 0.0021 for WT E12.75 vs bikunin −/− E12.75, n = 6 embryos for WT E12.25, n = 10 embryos for WT E12.5, n = 12 embryos for WT E12.75, n = 10 embryos for Tsg6 −/− E12.75, n = 6 embryos for bikunin −/− E12.75). Error bars represent mean ± SEM. See also Figure S7, S8, and Table S1. Scale bars: A (1000 µm); B (100 µm).
