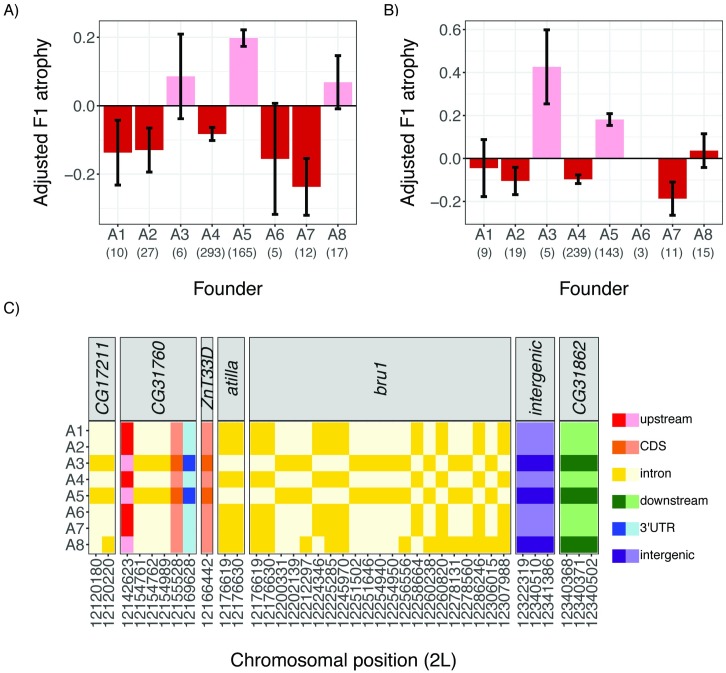
Fig 3. Phenotypic classes of founder alleles at the major QTL peak.
The mean adjusted F1 atrophy among 3-day-old females (A) and 21-day-old females (B) is shown for RILs carrying each of the eight founder alleles. Error bars denote standard error. QTL phasing (see Materials and methods) detects two allelic classes for both the 3-day-old and 21-day-old phenotypes: a sensitive allele that increases the odds of F1 ovarian atrophy (pink) and a tolerant allele that decreases the odds of F1 ovarian atrophy (red). The assignment of founder alleles to phenotypic classes across ages is consistent for all founders except A8. (C) In-phase SNPs in Population A RIL founder genomes, indicated by their position in the Drosophila melanogaster reference assembly (dm6, [45]). SNPs are colored according to the function of the affected sequence, and shaded according to whether the founder exhibits the reference (light) or alternate (dark) allele. The individual numerical values required to generate bar plots for panels A and B can be found in S4 and S5 Data, respectively. In-phase polymorphisms represented in panel C are provided in S1 Table. bru1, bruno CDS, coding sequence; dm6, Drosophila melanogaster reference assembly; F1,filial 1; QTL, quantitative trait locus; RIL, recombinant inbred line; SNP, single nucleotide polymorphism.