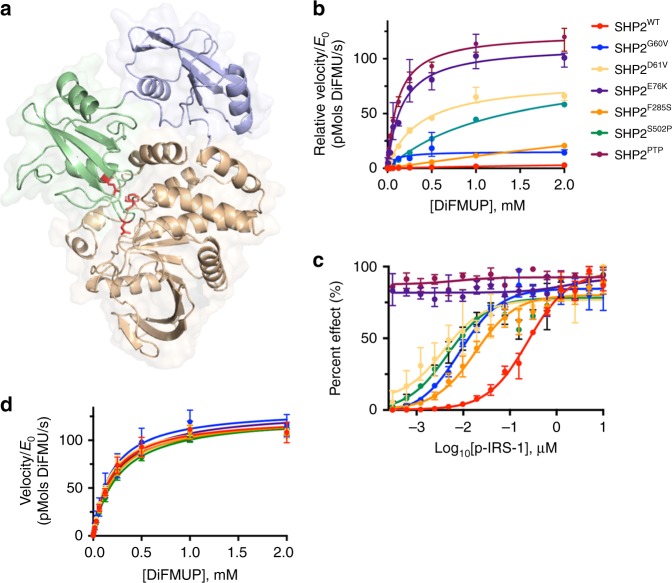
Fig. 3.
SHP2 cancer mutations enhance basal activity and response to phospholigands. a Sites of oncogenic mutations of SHP2 mapped onto the autoinhibited structure of SHP2 (PDB: 2SHP), which localize to the N-SH2:PTP interdomain interface. The N-SH2 domain of SHP2 is displayed in blue, C-SH2 in green, PTP in beige, and cancer mutations in red. b Normalized basal activity of SHP2WT and cancer mutants (5 nM WT, 1 nM F285S, 0.2 nM G60V, 0.1 nM D61V & S502P, and 0.05 nM E76K & PTP), plotted as a function of DiFMUP concentration. c Enzymatic activity of SHP2 analogs (0.05 nM) in the presence of DiFMUP (200 μM) plotted as a function of the concentration of a synthetic IRS-1-derived bisphosphorylated peptide (p-IRS-1, SLNY(p)IDLDLVK-dPEG8-LSTY(p)ASINFQK). d Enzymatic activity of various SHP2 proteins (0.05 nM) plotted as a function of DiFMUP concentration in the presence of saturating (10 μM) p-IRS-1. Data points are presented as mean ± standard deviation (n = 3)