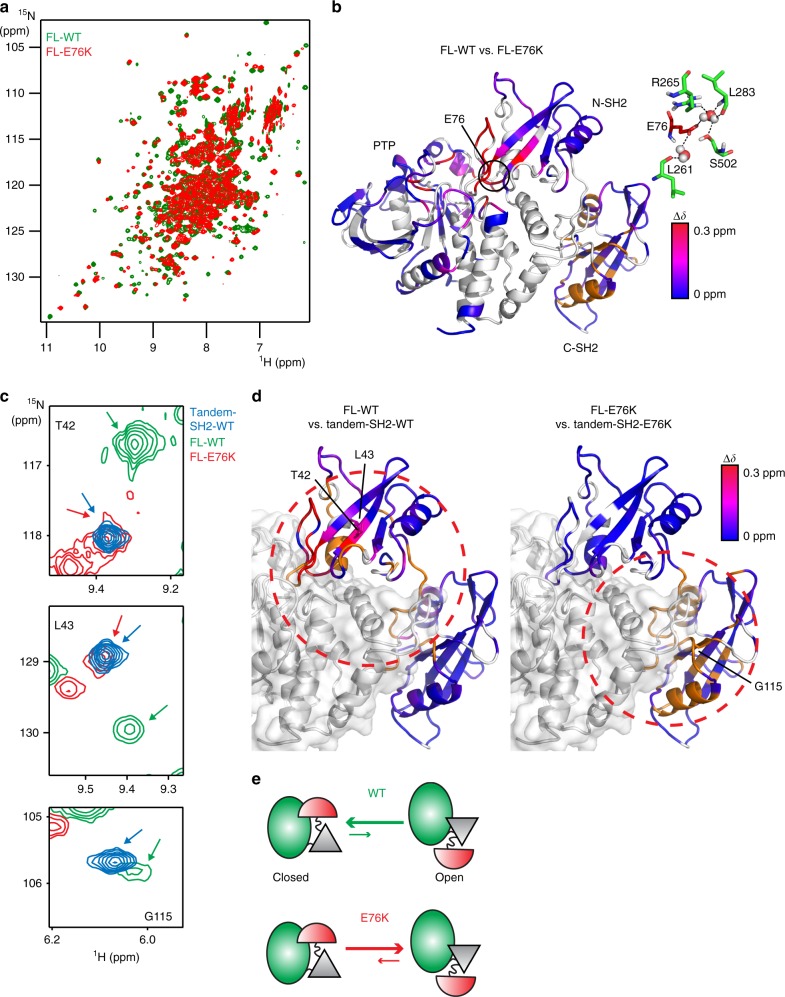
Fig. 1.
NMR chemical shift perturbations caused by the activating disease mutation E76K of SHP2. a Superposition of the [1H-15N]-TROSY-HSQC spectra of FL-WT and FL-E76K. b Chemical shift differences of a were plotted on the closed, inactive crystal structure of FL-WT (PDB 4dgp15). Unassigned, overlapping, or prolines residues are shown in gray; the orange color represents residues that experience chemical shift perturbations, but the precise value of Δδ is unknown because assignments are only available for one of the proteins. The interactions between E76 and residues in the PTP domain are shown on the right, with water molecules displayed as spheres. c Selected regions of the overlaid spectra of FL-WT, FL-E76K, and tandem-SH2-WT (see also Supplementary Fig. 2). d A red, dotted circle highlights major chemical shift perturbations in both SH2 domains for the isolated tandem-SH2 versus FL-WT (left), and only in the C-SH2 domain for the same comparison of E76K (right). Color coding is the same as in panel b. e Schematic representation of the open/closed conformational equilibrium of FL-WT and FL-E76K (N-SH2, red; C-SH2, gray; PTP, green)