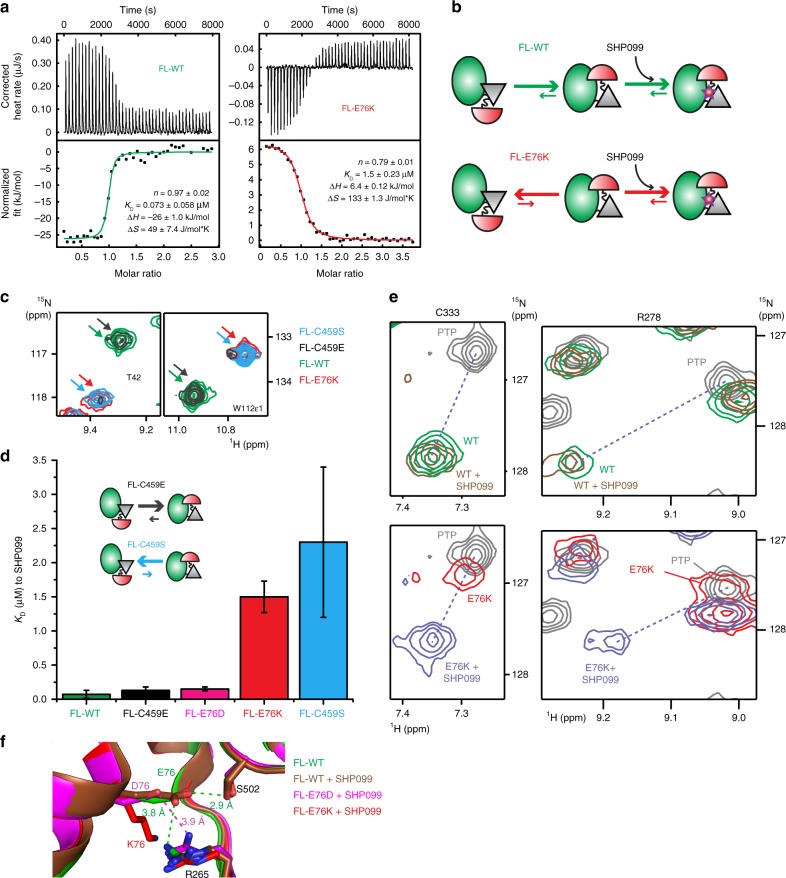
Fig. 5.
Comparing thermodynamics of SHP099 binding to wild-type and mutants of SHP2. a ITC profiles for the binding of SHP099 to FL-WT and FL-E76K (left, 30.5 μM of FL-WT titrated with 320 μM of SHP099; right, 82 μM of FL-E76K titrated with 800 μM of SHP099). b Scheme representing the proposed SHP099 binding mechanism with inverted equilibria between open and closed conformations as illustrated by size of kinetic arrows. c Selected regions of NMR spectra show that FL-C459E and FL-C459S resemble FL-WT and FL-E76K, respectively. d Bar graph summarizing the SHP099 dissociation constants determined by ITC for the denoted proteins (see also Supplementary Fig. 6e-f) together with a scheme for the open/close equilibrium of the dead mutants; uncertainties were derived from the fit to the data. e Chemical shift analysis to determine the population of the closed forms of FL-WT and FL-E76K. Dashed lines indicate the linear path between peaks in the PTP and FL-WT + SHP099 spectra. Using PTP and SHP099-bound chemical shifts for the completely open and closed states, respectively, the fraction of the closed state for FL-WT and FL-E76K are 0.90 and 0.04, respectively. f Superimposition of FL-WT (green), and protein/drug complexes FL-WT (brown), FL-E76D (magenta) and FL-E76K bound to SHP099 (red), reveals that the E76K mutation disrupts a hydrogen bond (E76–S502) and a salt bridge (E76–R265) between the N-SH2 and PTP domains, whereas the E76D mutation maintains the electrostatic interaction between D76 and R265