Figure 8.
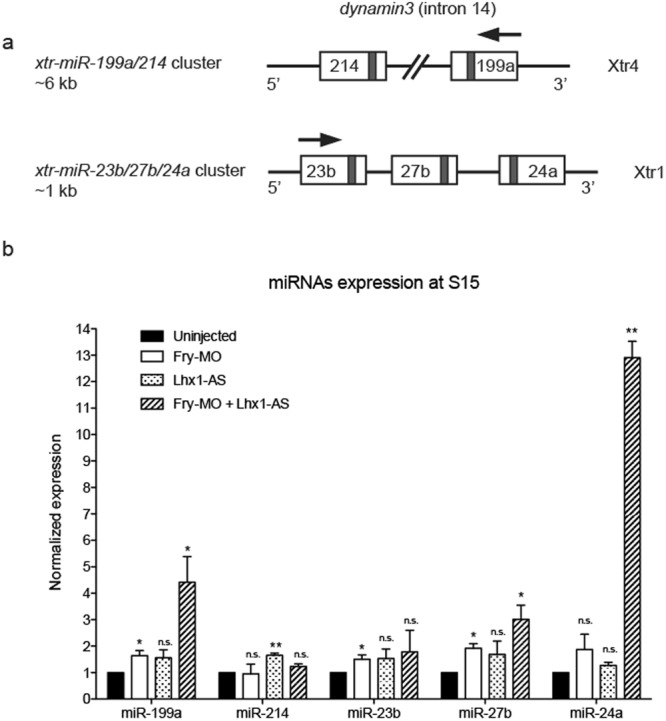
Validation of miRNA deep sequencing data by real-time qRT-PCR. (a) Schematic of the miR-199a/214 and miR-23b/27b/24a clusters in the X. tropicalis genome. The Xenopus tropicalis chromosome (Xtr) where each cluster is located is indicated. The miR-199a/214 cluster is located within intron 14 of the dynamin3 gene and has a length of ~6 kb while the miR-23b/27b/24a cluster is intergenic and has a length of ~1 kb. The grey colored rectangles indicate the position of the mature miRNAs within the pre-miRNA structures. Arrows indicate transcription direction. (b) Expression levels of miRs within the miR-199a/214 and miR-23b/27b/24a clusters were determined in Lhx1- and/or Fry-depleted embryos. 8-cell embryos were injected 2x V2 with 2.5 ng of Fry-MO and/or 50 pg of Lhx1-AS. Error bars indicate standard deviation derived from three repeats of the PCR reactions with different biological samples. Statistical significant differences were determined by a one-tailed paired t-test. n.s., non-significant; *p < 0.05; **p < 0.01.
