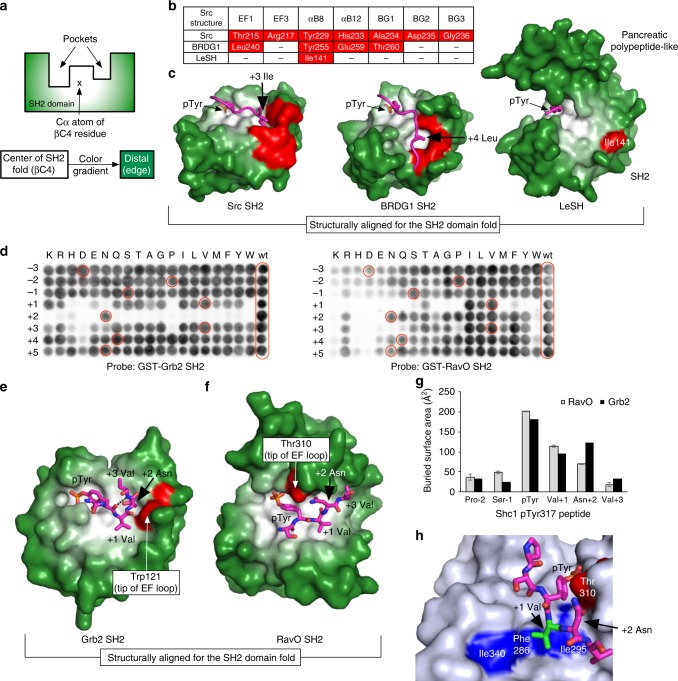
Fig. 6.
The Legionella SH2 domains lack a specificity pocket. a A color gradient depicting the distance of the ligand-binding pockets on a typical eukaryotic SH2 domain to the center of the globular SH2 domain fold. The Cα atom of the βC4 residue of an SH2 domain was defined as the origin of the color gradient, from white to dark green, to facilitate visualization of the specificity pocket in an SH2 domain. b Residues that contribute to forming the inside lining of either the P + 3 or + 4-binding pocket in SH2 domains. The P + 3 (Ile)-binding pocket in the Src SH2 domain involves the EF1, EF3, BG2, and BG3 residues whereas the P + 4 (Leu)-binding pocket in the BRDG1 SH2 domain is formed by the EF1, αB8, αB12, and BG1 residues. A 3D structural alignment of the two SH2 domains with LeSH was used to identify structurally equivalent residues in the latter. The symbol (–) indicates absence of an equivalent residue in the structure. c Comparison of the structure of the LeSH-DnaJ-A1 peptide complex with those of the Src and BRDG1 SH2 domains. The bound peptides are pTyr-Glu-Glu-Ile for the Src SH2 domain, and pTyr-Glu-Asn-Val-Leu for the BRDG1 SH2 domain. Only the side chains of the key residues (Ile + 3 for the Src SH2 ligand and Leu + 4 for the BRDG1 SH2 ligand) are shown in magenta sticks. The pocket-forming residues listed in b are colored red. d A permutation array based on the Shc1 pTyr317 peptide (DPSpYVNVQNL) probed with either the GST-fused Grb2 (left panel) or RavO SH2 domain (right panel). The amino acid at each position between −3 and +5 was replaced by one of 19 natural amino acids (except for Cys). The pTyr residue was fixed. The peptide spots identical to the wild-type (wt) sequence are highlighted in red circles. e, f Comparison of the LeSH and human Grb2 SH2 domains bound to the same peptide (Shc1 pTyr317). The human Grb2 SH2 domain (e) is structurally aligned with the L. pneumophila RavO SH2 domain (f). The Shc1 peptide Ser-pTyr-Val-Asn-Val is shown in magenta sticks. The residue located at the tip of the EF loop is colored red for each SH2 domain. g The buried surface area (in Å2) calculated for each residue of the Shc1 pTyr317 peptide bound to the SH2 domain of RavO or Grb2. The error bars for the area from the RavO complex indicate deviations between the three copies of SH2-peptide complexes in the asymmetric unit (Supplementary Fig 9e and f). h The hydrophobic patch on the RavO SH2 domain used for accommodating the +1 position of the ligand peptide