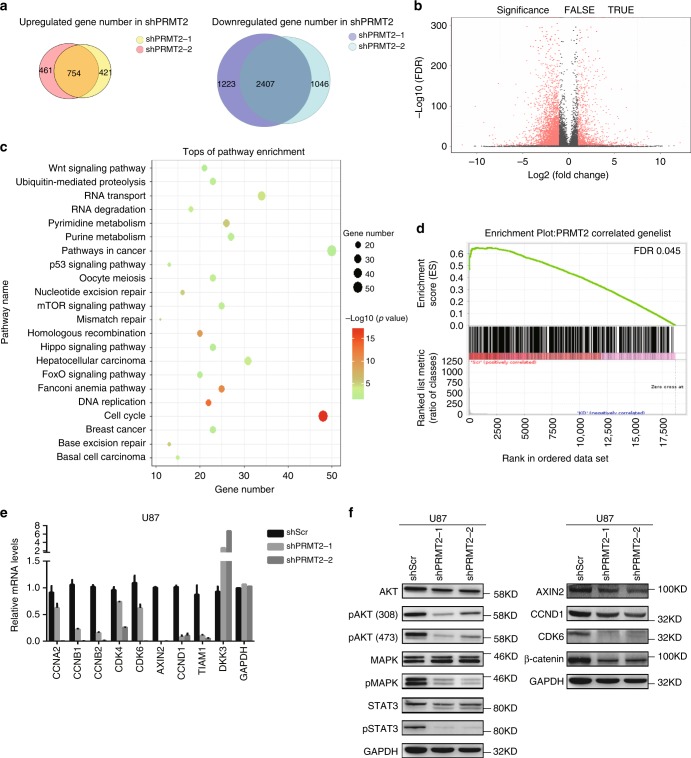
Fig. 4.
PRMT2 is essential for the activation of oncogenic transcriptional programs. a Venn diagrams show 754 upreglated genes with overlap in U87 shPRMT2-1 compared with shPRMT2-2 (top panel) and 2407 downregulated genes with overlap in U87 shPRMT2-1 compared with shPRMT2-2 (bottom panel). b Volcano plot shows the transcript levels differentially expressed between U87 shScr and U87 shPRMT2. Filtered by log2 (fold change) ≥ 1 and false discovery rate (FDR) < 0.001, significantly dysregulated genes are shown as red dots. The representative genes involved in cell cycle progression and pathways in cancer are labeled. Upregulated genes number = 1215, whereas downregulated genes number = 3630. c KEGG pathway enrichment analyses of significantly downregulated genes in U87 shPRMT2. Tops of enriched pathway are shown. d GSEA indicates PRMT2 positively correlated genes (583 genes, p < 0.001 and r > 0.3)) in the TCGA dataset is significantly enriched in the control group (Scr), compared with PRMT2-depleted group (KD). e RT-qPCR analysis of deregulated genes involved in cell cycle progression and pathways in cancer in U87 cells transduced with shScr or shPRMT2. Error bars ± SD, n = 2. f WB analyses evaluating cell cycle genes and oncogenic pathways in U87 cells expressing shScr or shPRMT2. GAPDH was used as loading control