FIG 1.
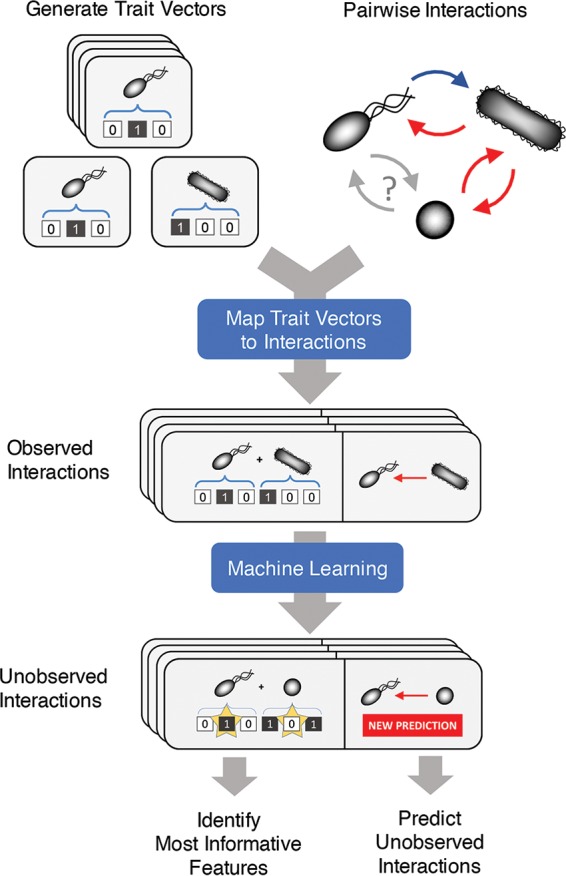
Schematic representation of our machine learning approach for inferring interactions among microbes. A trait vector captures the characteristics of each organism in the community of interest. The presence or absence of a trait in a given organism is encoded (as a binary number) in the corresponding element of the trait vector. For every possible pairwise interaction among community members, we construct a composite vector that is the concatenation of the corresponding trait vectors. The vector of the organism whose response is being predicted is concatenated to the front of the trait vector of its interaction partner. For the set of observed interactions, each composite vector is then mapped to the measured response of the interacting species. All observed interactions are then used to train a model that predicts the outcome of unobserved interactions. If random forest is used, then feature contributions can be calculated on a case-by-case basis to identify which elements of the composite genome contribute most strongly to the prediction.
