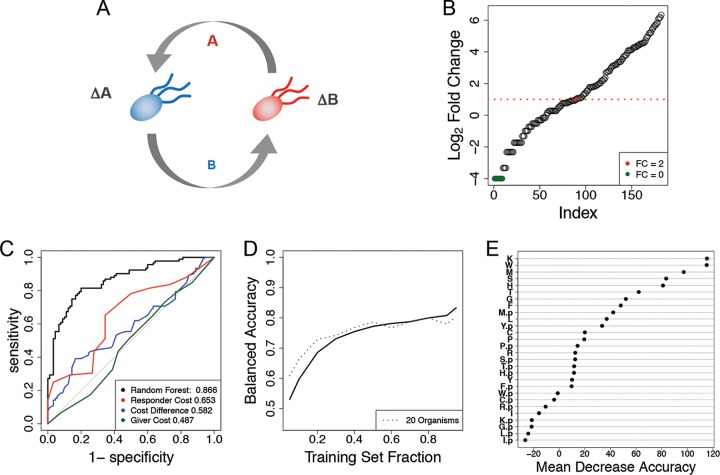
FIG 4.
Data representation and results for the case study of a network of auxotrophic E. coli strains. (A) In the original experiment, single-gene knockout E. coli auxotrophs were cocultured in minimal medium. For the ΔA mutant to grow, it must receive amino acid A from the ΔB mutant, which in turn must receive another amino acid, B, for growth. Auxotroph strains were constructed for the following amino acids: cysteine, phenylalanine, glycine, histidine, isoleucine, leucine, methionine, proline, arginine, serine, threonine, tryptophan, and tyrosine. (B) Auxotroph strain fold changes in ascending order. E. coli strains had a weak response (fold change ≤ 2) 90 times and failed to grow 9 times (green circles). In 92 instances, the E. coli auxotroph population more than doubled over the course of 84 h. (C) ROC curves for all 182 observations were determined for a random forest classifier using 28 amino acids as predictors. Single-value thresholds based on the biosynthetic costs of knocked-out amino acids resulted in poorer performance than the random forest algorithm. (D) Trajectory of a learning curve built for the E. coli interactions (solid line) closely resembles that of the learning curve for in silico communities with 20 organisms (dashed line). (E) Ranking of 28 amino acids according to their effects on prediction accuracy when randomly permuted. Amino acids corresponding to the receiver strain are enriched near the top of the list. A suffix “p” indicates that the predictive feature belongs to the giver strain. A single case of a Δmethionine mutant cocultured with a Δcysteine mutant is shown in Fig. S2.