Fig. 2.
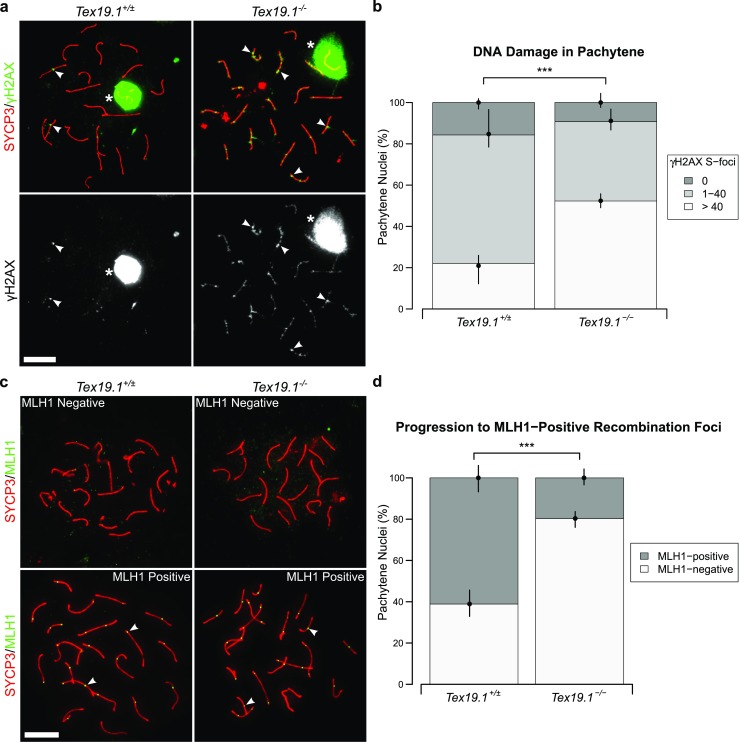
Autosomally synapsed pachytene Tex19.1−/− spermatocytes have skewed recombination profiles. a Immunostaining for γH2AX (green) in Tex19.1+/± and Tex19.1−/− autosomally synapsed pachytene spermatocyte chromosome spreads. SYCP3 (red) marks the lateral element of the synaptonemal complex. A single channel greyscale image for γH2AX is also shown. The Tex19.1+/± image belongs to the intermediate γH2AX foci category (1–40 S-foci), the Tex19.1−/− image belongs to the high γH2AX foci category (> 40 S-foci). Asterisks indicate sex bodies, and arrowheads indicate example S-foci associated with axes. Scale bars 10 μm. b Quantification of γH2AX S-foci; 172 and 120 autosomally synapsed Tex19.1+/± and Tex19.1−/− pachytene nuclei were categorised according to the number of γH2AX S-foci; 22.1, 62.2 and 15.7% of pachytene Tex19.1+/± spermatocytes have > 40, 1–40 and 0 γH2AX foci, respectively, compared to 52.3, 38.5 and 9.2% Tex19.1−/− pachytene nuclei. Asterisks denote significance (χ2 test, ***p < 0.001). Nuclei were obtained from three animals for each genotype, and circles and vertical lines represent the means and interquartile distances of animals within each genotype. c Immunostaining for MLH1 (green) in control and Tex19.1−/− autosomally synapsed pachytene spermatocyte chromosome spreads. SYCP3 (red) marks the lateral element of the synaptonemal complex. Examples of MLH1-positive and MLH1-negative images are shown. Nuclei containing ten or more axis-associated MLH1-foci (arrowheads) were classified as MLH1-positive. Scale bars 10 μm. d Quantification of pachytene nuclei that are MLH1-positive; 162 autosomally synapsed pachytene nuclei were scored for each genotype; 61.1% Tex19.1+/± and 19.8% Tex19.1−/− nuclei were MLH1-positive. Asterisks denote significance (χ2 test, ***p < 0.001). Nuclei were obtained from three animals for each genotype, and circles and vertical lines represent the means and interquartile distances of animals within each genotype
