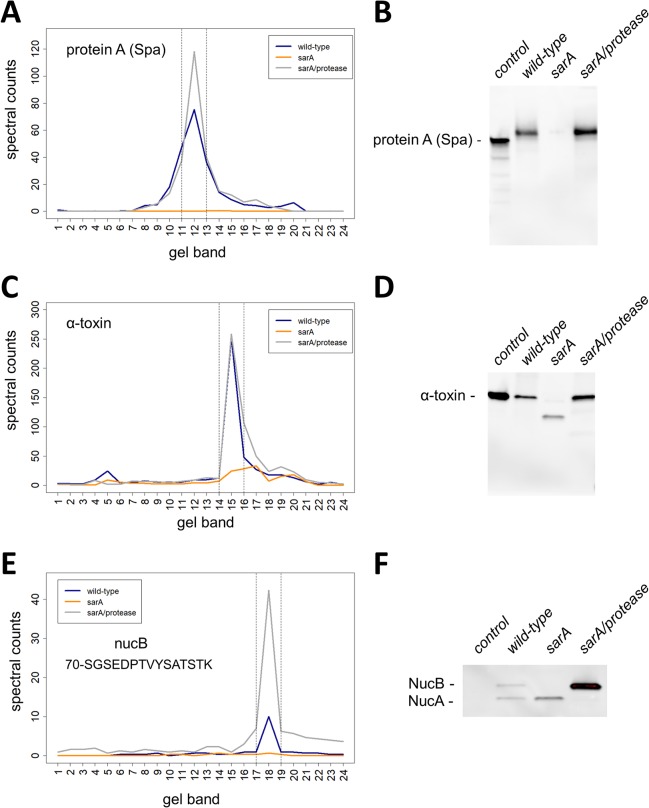
Figure 3.
Validation of exoproteins decreased in sarA-mediated extracellular protease-dependent and -independent mechanisms. The amounts and mass distributions of Spa (A), alpha-toxin (C), and NucB (E) were visualized by plotting the total spectral counts averaged across three biological replicates as a function of gel band. Spectral counts for the single unique NucB peptide (70-SGSEDPTVYSATSTK) were used for panel E. The vertical lines indicate the 3-band window used for data analysis in the full-length protein method. Immunoblot for Spa (B), alpha-toxin (D), and NucB (F) verified the respective spectral counting data. Shown is a representative immunoblot from biological triplicates. Controls for immunoblots are purified Spa that is known to migrate at slightly lower molecular mass relative to in vivo expressed Spa (B), purified alpha-toxin protein (D), and a control strain that does not express NucB (F). NucA is a proteolytically processed version of NucB (F).