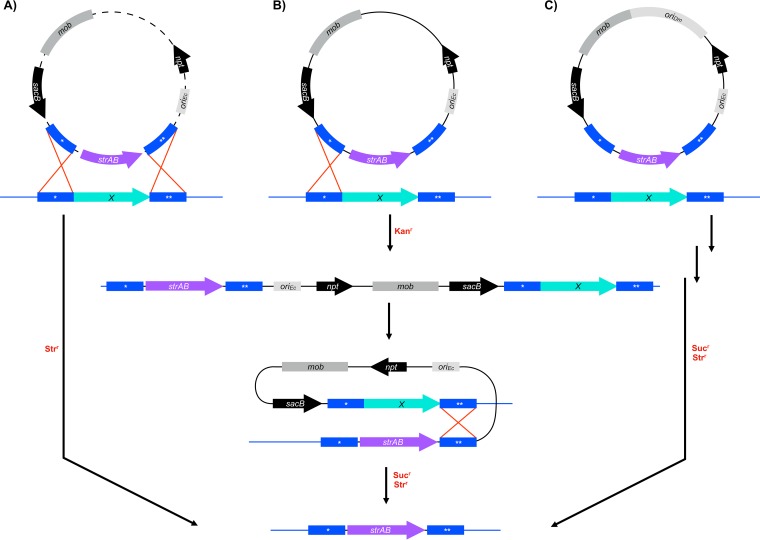
FIG 1.
Schematic of deletion methods used in Desulfovibrio spp. Plasmids (black lines) were designed to replace a target gene (X, aqua arrows) in the chromosome (blue lines) with a streptomycin resistance cassette (strAB, purple arrows). Regions upstream (*) and downstream (**) of the target gene (blue boxes) on the chromosome undergo recombination (red lines) with homologous regions that are cloned into the deletion plasmid. Key steps, such as recombination events (red crosses), are indicated in the boxes, and the selection steps are labeled in red. (A) Double recombination can occur in one step after plasmids are linearized (dashed lines) by endogenous restriction enzymes. Mutants are selected using the marker (e.g., strAB) that was exchanged with the target gene. (B) Two-step double recombination is possible when suicide vectors integrate into the chromosome in the first homologous recombination event and then recombine out after the second homologous recombination event. The first step and second step are selected for with antibiotic resistance markers (e.g., npt) and counterselectable markers (e.g., sacB), respectively. (C) A replicative deletion plasmid designed to target genes for deletion may undergo double recombination in one or two steps as shown in panels A and B, respectively. After passaging the cells without antibiotic, the mutants are selected with an antibiotic resistance cassette (e.g., strAB) and a counterselectable marker (e.g., sacB). mob, mobilization genes (mobA′, mobB, mobC); npt, kanamycin-resistance gene; oriDm, origin of replication for D. magneticus; oriEc, origin of replication for E. coli.