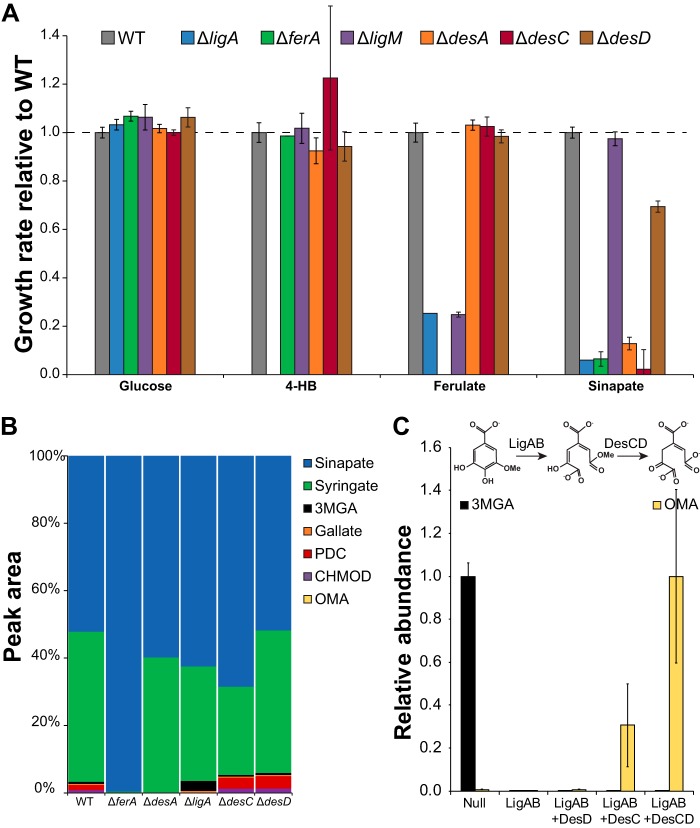
FIG 6.
Gene deletions identify enzymes for sinapate degradation. (A) Mutant strains with clean deletions of the indicated genes were grown in minimal medium containing the indicated carbon source. Disruptions of the newly identified genes desC and desD decrease growth with sinapate but not with any of the other substrates tested. Error bars show one standard deviation, calculated from three biological replicates. (B) Mutant strains were grown in minimal medium containing glucose and sinapate. Accumulation of downstream metabolites was monitored by LC-MS. Results show the averages from two technical and three biological replicates. (C) Crude lysates expressing the indicated enzymes were combined and incubated with 3MGA. Conversion was monitored by LC-MS. Abundance is calculated relative to the highest measurement for a given compound. Error bars show the standard deviations from three biological replicates.