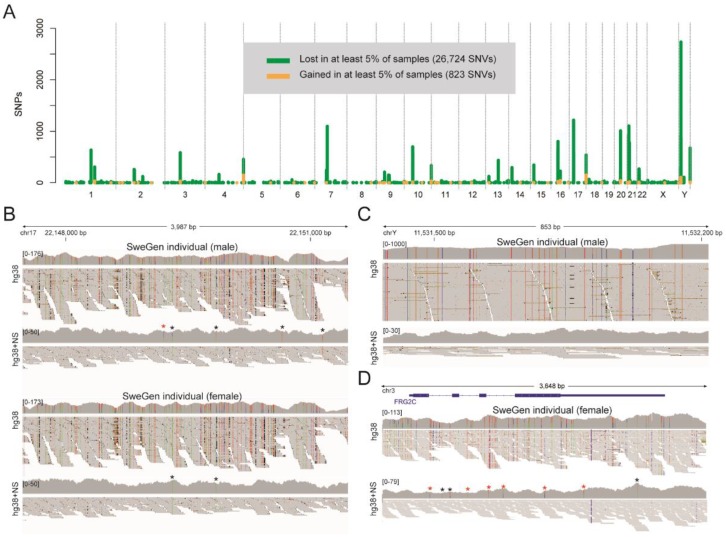
Figure 5.
A novel reference gives improved alignment and SNV calling of SweGen WGS data. (A) Genomic distribution of SNVs that are lost (green) and gained (orange) when NS are appended to the hg38 reference. Only non-centromeric SNVs that are lost/gained in at least 5% of the 200 SweGen samples are shown in this figure. (B) An IGV [31] view of Illumina reads for two representative SweGen samples at a region on chr17, where some SNVs are lost and others are gained when using the hg38+NS reference. Illumina data is shown for a male and a female (not the same individuals as Swe1 and Swe2). Both for the male and female, the coverage decreases over the region when NS are appended to hg38, and about 100 (homozygous) false positive SNV calls are lost in each of the samples. Only five heterozygous SNVs where found for the male individual when the novel reference was used, and two homozyogous SNVs for the female (marked by asterisks ‘*’). A red asterisk indicates a gained SNV that is not detected in hg38. (C) An example region on chrY where the coverage was reduced from almost 1000× to below 30× when using hg38+NS, and where a large number of SNVs were lost. Only data for the male individual is shown in this panel. (D) Improved alignment and SNV calling over the FRG2C locus on chromosome 3. A large number of SNVs were lost, and six SNVs were gained (red asterisks ‘*’), in the female SweGen sample. Some of the lost and gained SNVs are located in the coding sequences of FRG2C.