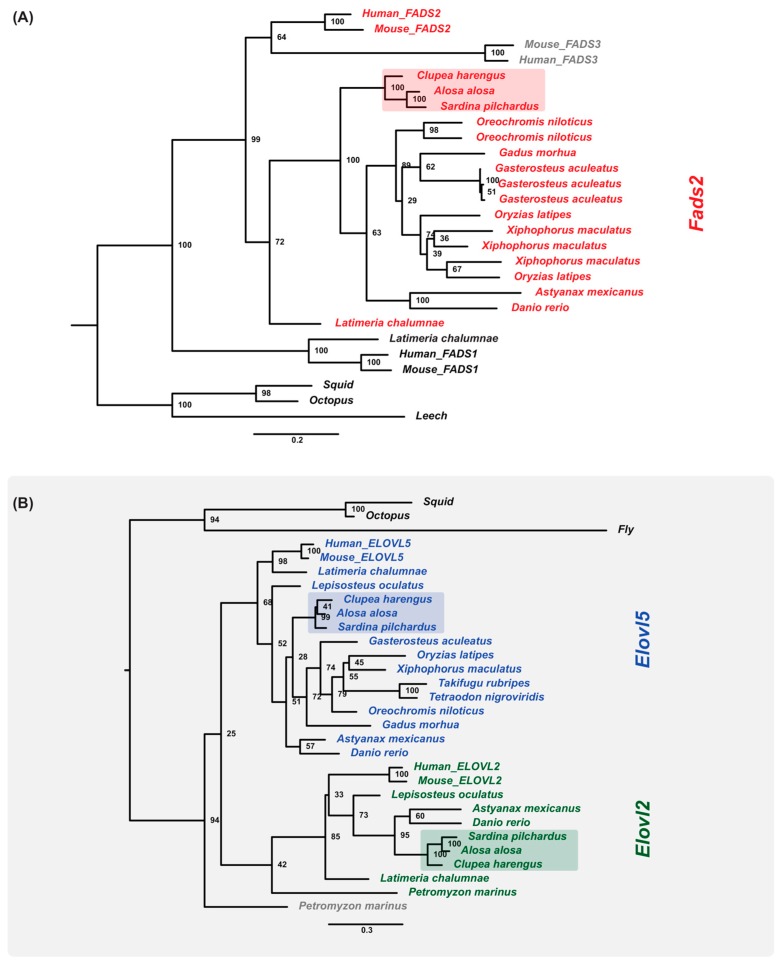
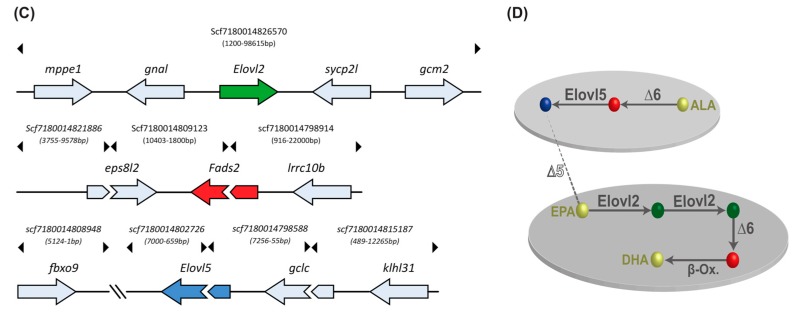
Figure 3.
Maximum likelihood phylogenetic analysis of fads2 (A) and elovl orthologues (B) analyzed in the present study: Clupeiformes species are highlighted, node numbers indicate bootstrap values. (C) Reconstructed genomic loci of fads2, elovl2, and elovl5 denote synteny conservation between the European sardine and Atlantic herring: scaffold coordinates and identified neighbouring genes are indicated; broken lines and arrows denote reconstruction from overlapping scaffolds. (D) LC-PUFA biosynthesis pathway in the European sardine, dashed line indicates the Δ5 desaturation capacity absent in the European sardine, n-3 fatty acids are indicated in yellow: ALA—α-Linolenic acid (18:3n-3), EPA—eicosapentaenoic acid (20:5n-3) and DHA—docosahexaenoic acid (22:6n-3).