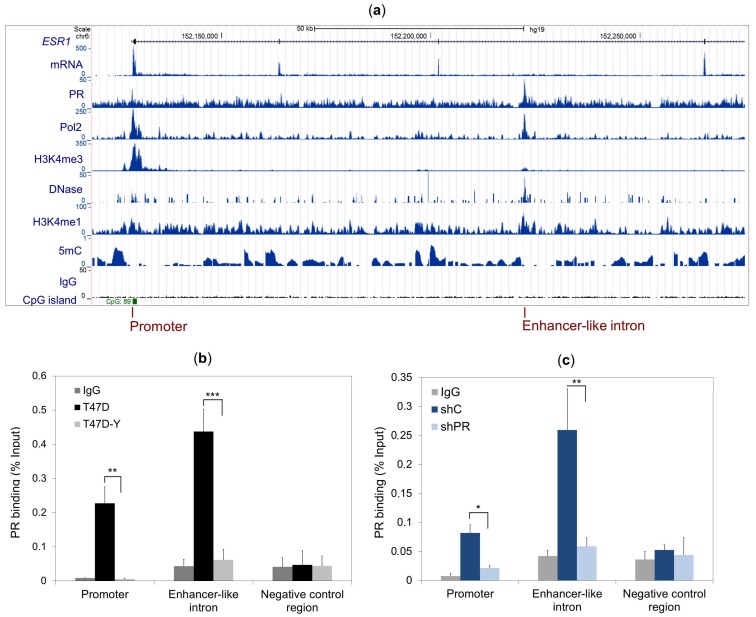
Figure 2.
PR binds to the promoter and to an enhancer-like intron of the ESR1 gene in hormone-deprived T47D breast cancer cells. (a) Screen shot from the UCSC genome browser showing the ESR1 gene, the RNA reads and the ChIP-seq results from PR binding, with a peak at the gene promoter marked by polymerase 2 binding (Pol2), histone 3 trimethylated at lysine 4 (H3K4me3), low DNA methylation (5 mC) and a CpG island at the bottom. Another PR peak is found in an intronic region containing the classical enhancer epigenetic marks of the DNase hypersensitive site (DNase), histone 3 monomethylated at lysine 4 (H3K4me1) and low DNA methylation signal (5mC). The negative control immunoprecipitation is indicated by the IgG antibody. (b,c) The ChIP assay was performed with a specific antibody against PR or total rabbit IgG in T47D cells and PR-deficient cells (T47D-Y) (b) or PR-depleted cells (shPR, clone trcn0000010776) and control cells expressing a scrambled shRNA (shC) (c). Specific binding was assessed by quantitative PCR amplification of the ESR1 gene promoter, an enhancer-like intronic sequence and a genomic region localized at the 3′-end of the enhancer-like intron (negative control region). Error bars represent the SD of three independent experiments. * p less than or equal to 0.05, ** p less than or equal to 0.01, *** p less than or equal to 0.005, unpaired two-tailed Student’s t-test.