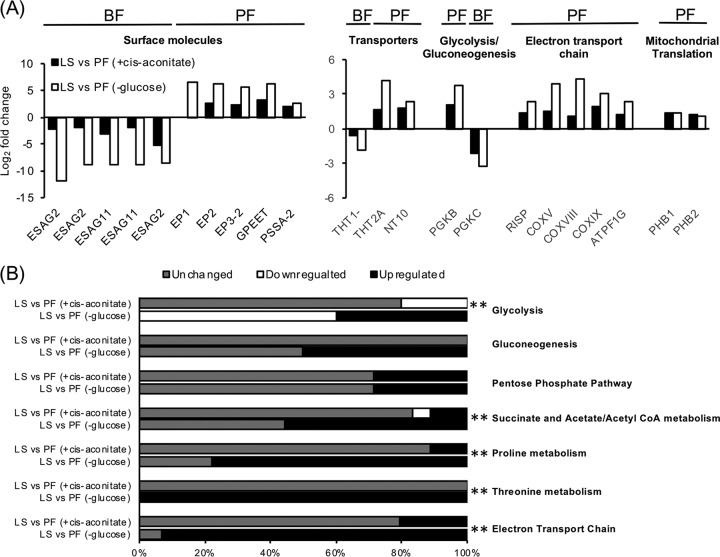
FIG 7.
Evaluation of pleomorphic PF differentiated by different cues at the transcriptome level. (A) A comparison of the relative transcript abundance of PF parasites to LS parasites for a subsect of life-stage-enriched genes. PF were generated by differentiation of SS parasites using either glucose-depleted medium (-glc) or cis-aconitate treatment (+cis-aconitate). All transcripts included had >2-fold change and adjusted P values < 0.05 except for THT1- in PF (+cis-aconitate), which was downregulated less than 2-fold with an adjusted P value > 0.05. N/A, data not available. ESAG2, expression site-associated gene 2; ESAG11, expression site-associated gene 11; EP1, EP1 procyclin; EP2, EP2 procyclin; EP3-2, EP3-2 procyclin; GPEET, GPEET procyclin; PSSA-2, procyclic form surface phosphoprotein; THT1-, glucose transporter 1B; THT2A, glucose transporter 2A; NT10, purine nucleoside transporter 10; PGKB, phosphoglycerate kinase B; PGKC, phosphoglycerate kinase C; RISP, Rieske iron-sulfur protein; COXV, cytochrome oxidase subunit V; COXVIII, cytochrome oxidase subunit VIII; COXIX, cytochrome oxidase subunit IX; ATPF1G, ATP synthase F1 subunit gamma protein; PHB1, prohibitin 1, PHB2, prohibitin 2. (B) The percentage of transcripts in major metabolic pathways (as identified by GO term definition and confirmed in TrypanoCyc [60]) that are regulated in PF differentiated by cis-aconitate or glucose depletion compared to LS. For glycolysis and gluconeogenesis, only enzymes that participate in one of the reactions were included. Upregulated transcripts are those that increased >2-fold and had adjusted P values < 0.05 in PF compared to LS, while downregulated transcripts were decreased >2-fold with adjusted P values < 0.05. Chi-squared goodness-of-fit tests were performed for genes in each pathway to determine whether the proportions of regulated genes between PF differentiated by cis-aconitate and glucose depletion were different. ** indicates P value < 0.01, suggesting transcripts involved in the pathway are differentially regulated in PFs differentiated by the two cues.