Fig. 1 |. Optimized workflow and experimental design of global proteome and phosphoproteome analysis in tissues using TMT.
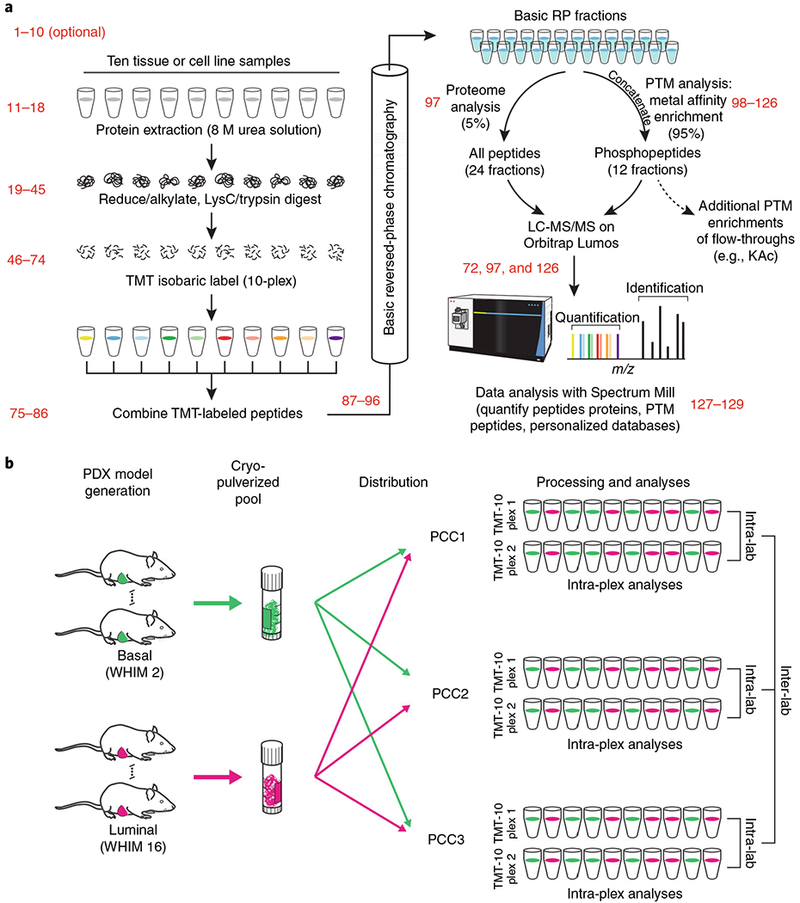
a, Multiple aspects of sample handling were optimized based on a preexisting workflow for global proteome and phosphoproteome analysis (Mertins et al.2). Some of the conditions tested relative to the preexisting workflow were (i) digestion at higher protein concentrations, which effectively increases the enzyme concentration during digestion, resulting in lower missed cleavage rates; (ii) reconstitution of lysyl endopeptidase in water, instead of 50 mM acetic acid, which better maintains the activity of the enzyme; (iii) quantification of peptides by BCA before isobaric labeling, which yields more accurate input amounts than BCA at the protein level; (iv) offline basic RP fractionation using either Agilent or Waters columns, which yield equivalent results; and (v) optimization of HCD energy for each individual instrument, rather than the use of a common collision energy, which improved spectral quality. The relevant steps of the Procedure are indicated in red. b, Multiple mice of basal (WHIM 2) and luminal (WHIM16) subtypes were grown, and the tumors of each subtype were pooled together. Tumors of each subtype from multiple mice were cryofractured and aliquots of the homogenized powders were distributed to the three different laboratories for global proteome and phosphoproteome analysis. Each laboratory analyzed 2× TMT-10 plexes. Intra-plex, intra-lab, and inter-lab comparisons were conducted to test depth of coverage and reproducibility. PCC1–3 indicate Protein Characterization Centers 1 (Broad Institute), 2 (Johns Hopkins University), and 3 (Pacific Northwest National Laboratory), respectively. BCA, bicinchoninic acid; HCD, higher-energy collision dissociation. a Adapted from Extended Data Fig. 1 in ref. 2, Springer Nature.
