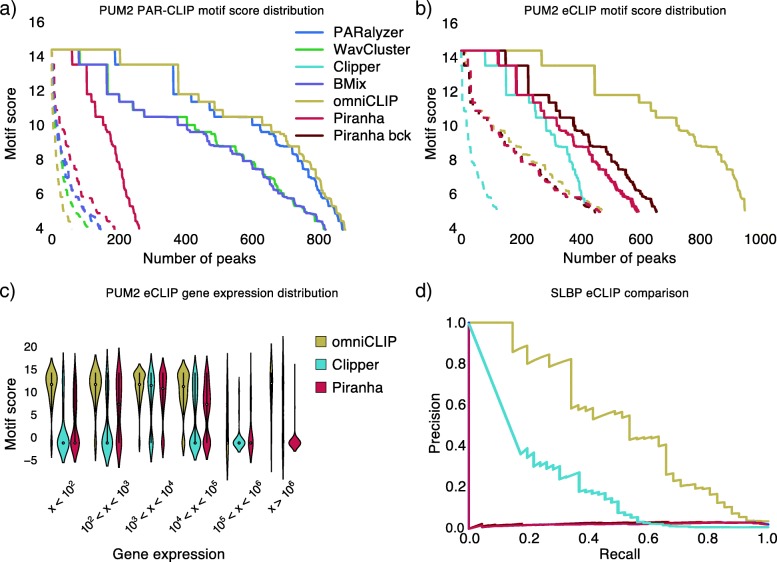
Fig. 3.
Performance evaluation. a Sorted distribution the PUM2 motif scores of the top 1,000 called peaks for PARalyzer, Piranha, WavCluster, BMix and omniCLIP on a HEK293 PUM2 PAR-CLIP dataset. Dashed lines indicate motif enrichments in a random control b PUM2 motif scores distribution for the top 1,000 called peaks for omniCLIP, Clipper and Piranha on a HepG2 PUM2 eCLIP dataset. c Motif score distribution for the top 1,000 peaks on the HepG2 PUM2 eCLIP dataset. Peaks are further classified by gene expression. d Precision recall curves for Clipper Piranha and omniCLIP on a HepG2 SLBP dataset for discriminating histone genes and non-histone genes from peak scores