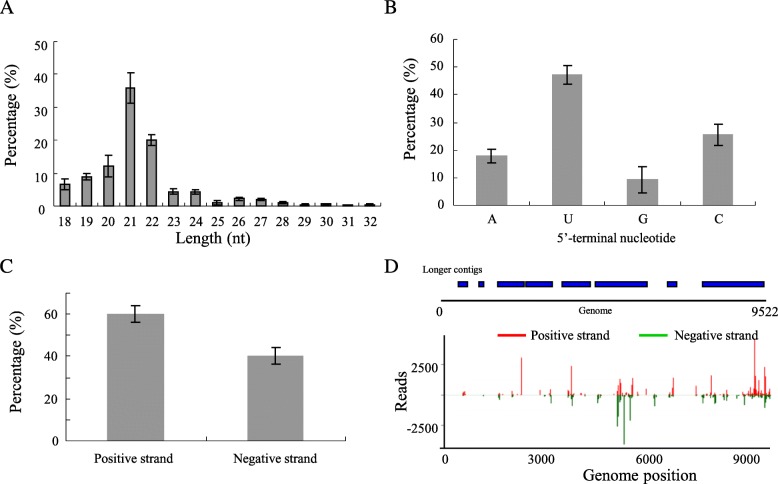
Fig. 2.
Characterizations of siRNAs (18–32 nt) derived from TeMV in P. edulis fruits through small RNA deep sequencing platform. a, Size distribution of siRNAs matching to TeMV in infected P. edulis fruits. b, Nucleotide bias of 5′-terminal nucleotide of siRNAs matching to TeMV in infected P. edulis fruits. c, Polarity distribution of siRNAs matching to TeMV in infected P. edulis fruits. “+” and “-” indicate siRNAs derived from positive and negative genomic strands, respectively. d, Coverage (up panel) and profile (down panel) of siRNAs along the reference TeMV genome in infected P. edulis fruits. a, b, c and d; values are mean of three independent experiments