Fig. 1.
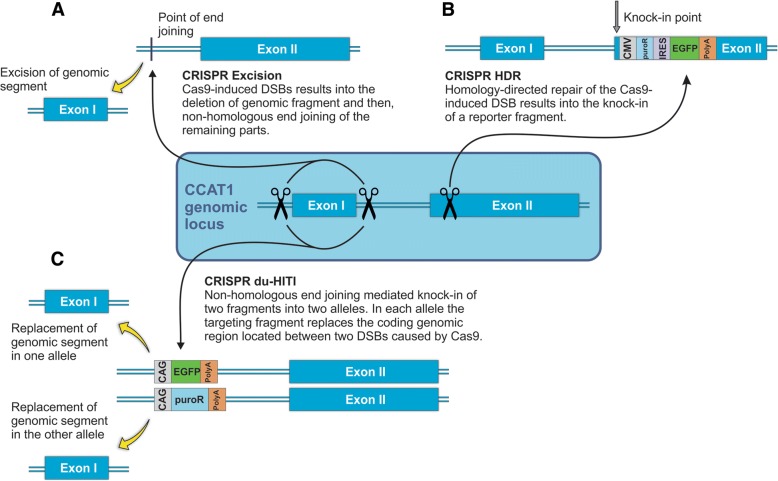
CRISPR/Cas9 knockout strategies for ablation of CCAT1 lncRNA gene. a “CRISPR excision”. To delete a genomic fragment (here, exon 1) two sgRNAs are targetted to either side of the fragment. Non- homologous end joining of the two remaining parts of genomic DNA after Cas9-induced double-strand breaks (DSBs) results in the deletion of the genomic fragment. b “CRISPR HDR”. In this strategy, using one sgRNA and Cas9-induced DSB in one region is followed by homology-directed repair using a reporter (CMV-PuroR-IRES2-EGFP) plus polyadenylation signal fragment (originated from a donor vector with homology arms). In this case, any transcript initiated from the first or second exon is confronted by a premature transcription termination. c “CRISPR du-HITI”. This strategy uses two donor vectors without homology arms. Two vectors containing sgRNA+PAM are used as donors, one with EGFP expression cassette, and the other with a PuroR expression cassette. Use of two sgRNAs directs the Cas9 protein towards the two either end of exon 1 at both alleles. Endonuclease function of Cas9 results into deletion of a genomic fragment (here, exon 1) from each allele, and linearization of two donor vectors. Selection of cells for their green color and their resistance to puromycin dihydrochloride results into cells with their both alleles targetted and knocked out