Figure 1.
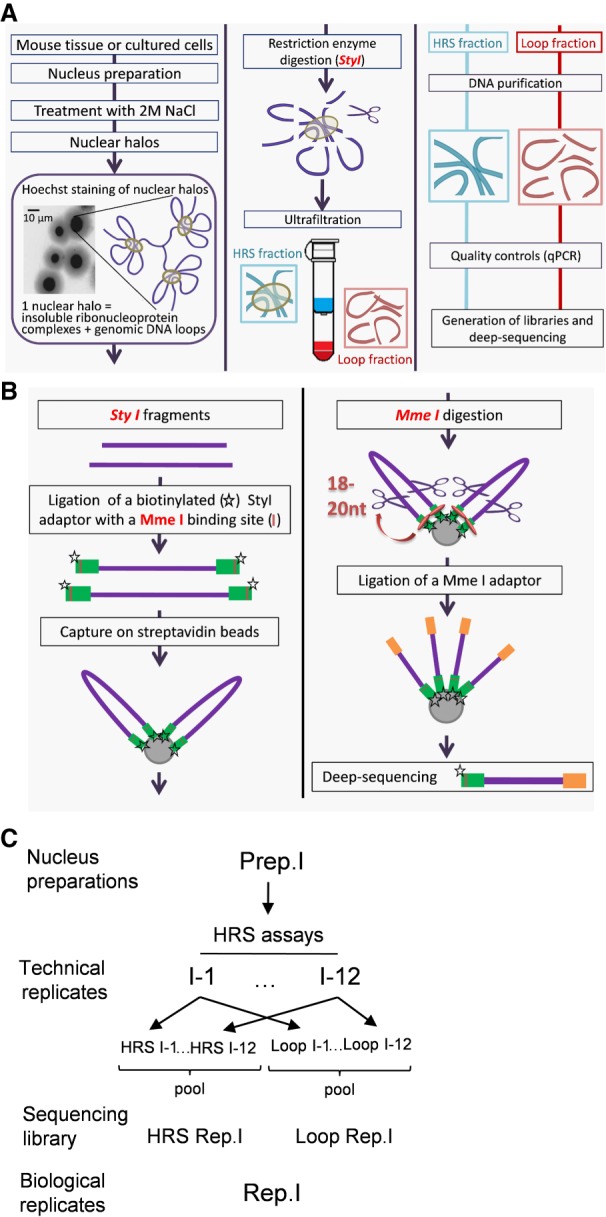
Flowchart of the HRS-seq method. The HRS-seq method consists of high-throughput sequencing of genomic DNA obtained from HRS assays. (A) HRS assay principle. Each HRS assay involves 105 nuclei that are treated with a 2 M NaCl buffer to obtain the so-called nuclear halos. Nuclear halos are digested with a restriction enzyme (here StyI), and the insolubilized fraction (HRS-containing fraction) is separated from the soluble Loop fraction by ultrafiltration. Genomic DNA is purified from each fraction, and controls are performed to ensure the quality of each assay. (B) Construction of HRS-seq libraries for deep-sequencing. For each HRS-seq experiment, two sequencing libraries are prepared: one from the HRS-containing fraction and one from the Loop fraction. A biotinylated StyI adaptor containing a MmeI binding site (green bars) is ligated to the StyI restriction fragments. Ligated fragments are captured on streptavidin beads and digested with MmeI to obtain StyI fragments having homogenous sizes (18 to 20 nt). The beads are washed several times, a MmeI adaptor (orange bars) is ligated, and these StyI/MmeI fragments are eluted from the beads. The StyI and MmeI adaptors are used for deep-sequencing. (C) Preparation of biological replicates. Each biological replicate (here Rep.1) is prepared from a different nuclei preparation (here Prep.1). A first sequencing library (here HRS Rep.1) is prepared from HRS fractions pooled from 12 HRS assays (technical replicates), and another one (Loop Rep.1) is prepared from Loop fractions pooled from the same 12 HRS assays. This procedure was applied to three distinct nuclei preparations to obtain six sequencing libraries representing three biological replicates.
