Figure 1.
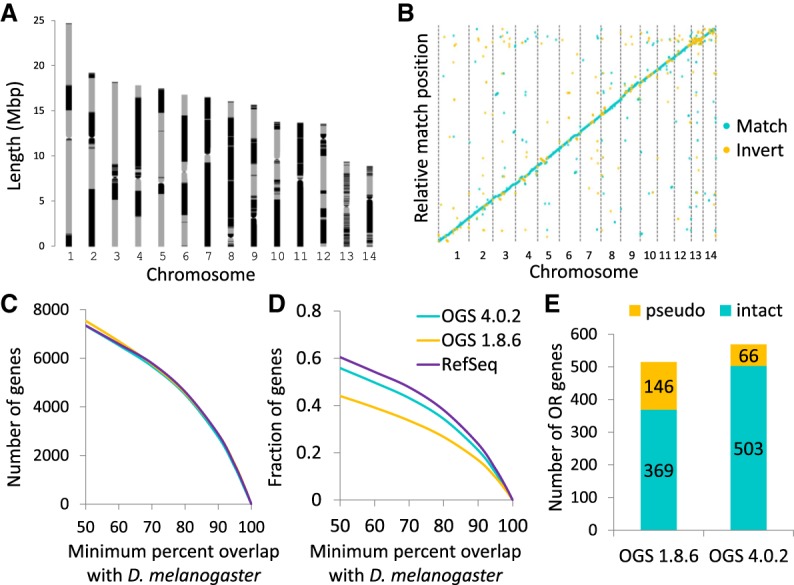
Genome assembly and annotation of the clonal raider ant. (A) Map of contigs within clonal raider ant chromosomes. Alternating gray and black indicate different contigs. Large stretches of uninterrupted gray or black indicate very large contigs, often containing most of a chromosome arm. Putative centromeres were identified based on Hi-C interaction (see Methods). (B) Whole-genome alignment of the second generation draft assembly (Oxley et al. 2014) with the new assembly. Largely continuous, noninverted alignment across the genome indicates little large-scale structural variation between the two assemblies. (C,D) BLAST hit overlap between clonal raider ant gene sets and the Drosophila melanogaster official gene set (v6). The good performance of OGS 4.0.2 in terms of number of genes, and intermediate performance in terms of fraction of genes, indicates high sensitivity and intermediate specificity. (E) Number of odorant receptors found in the annotation based on the previous assembly (OGS 1.8.6) vs. the current annotation (OGS 4.0.2). The large increase in intact ORs and decrease in predicted pseudogenes shows that the previous OR annotations were compromised by poor assembly and misassembly.
