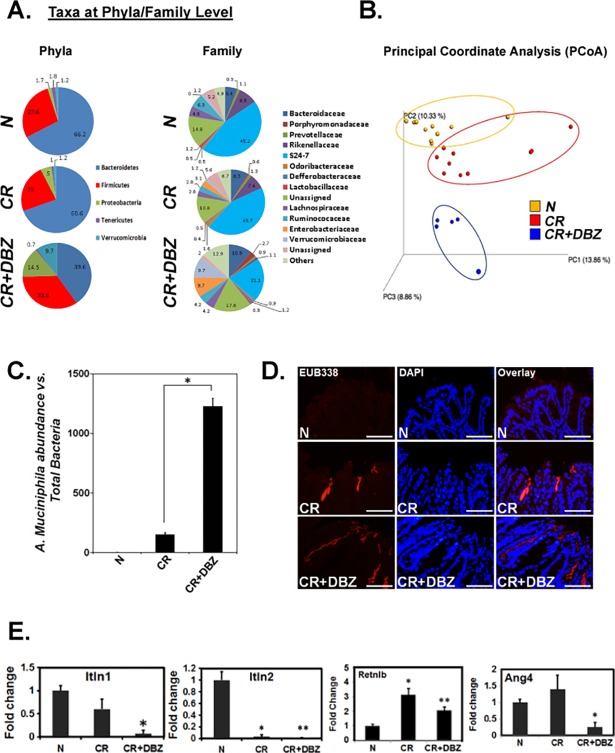
Fig 3. Evidence of bacterial dysbiosis during chronic blockade of Notch pathway in the colons of outbred mice.
A. Comparison of major microbial populations in fecal samples. Fecal samples from uninfected (N), CR-infected (CR) and CR+DBZ-treated mice were subjected to 16S rDNA sequencing and relative abundance of phyla and families were compared. Each chart represents the taxonomic composition in the indicated groups (n = 10 mice/group). B. Principal Coordinate Analysis (PCoA) of fecal microbiome composition. While N (shown in “orange”) and CR (shown in “red”) samples grouped closer to each other, CR+DBZ samples (shown in “blue”) shifted towards the opposite ends of the coordinates revealing distinct microbial communities (p<0.001; n = 10 mice/group). C. Species identification as a potential etiologic agent. Real-time qPCR showing relative abundance for A. mucinophila in various treatment groups (p<0.005; n = 3 independent experiments). D. Fluorescence microscopy to detect bacterial invasion. The attached bacteria in the flushed colonic tissues of N, CR or CR+DBZ mice were detected by FISH using a general bacterial 16S probe (TexasRed-Eub338; Bar = 100μm; n = 10 mice/group). DAPI was used as counter-stain. E. Effect of chronic Notch inhibition on anti-bacterial peptide gene expression. qPCR to examine expression levels of antibacterial peptide genes Itln1/2, Retnlb and Ang4 encoding Intelectin-1/2, Resistin-like molecule-β and Angiogenin-4, respectively (*, **p<0.005; n = 3 independent experiments).