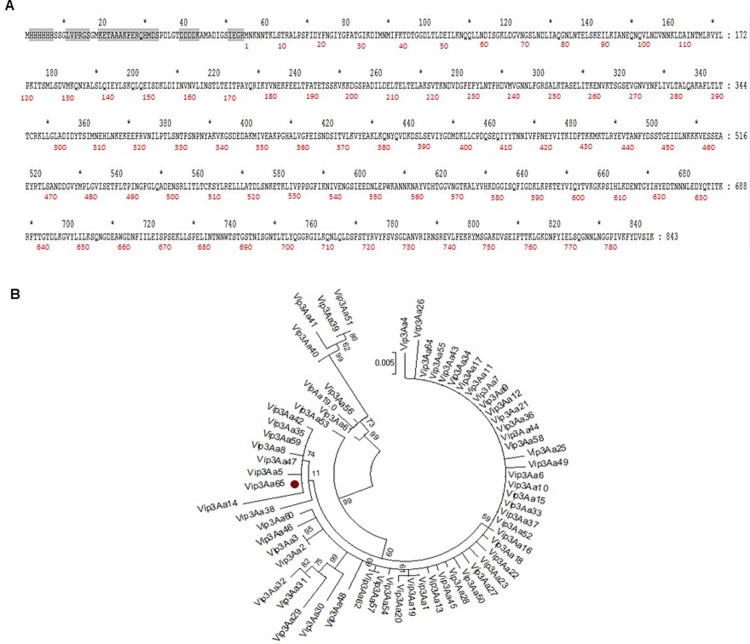
Fig 3. Vip3Aa65 amino acid sequence and phylogenetic tree of Vip3Aa proteins.
A) Assembled contig sequence of the Vip3Aa65 expressed protein construct. Gray boxes indicate the sequences for the His-tag, thrombin cleavage site, S-tag, enterokinase cleavage site, and Factor Xa cleavage site. Red numbers below the sequence refer to the amino acid sequence of the Vip3Aa65 protein (from amino acid 1 to 789). B) Maximum likehood tree of the Vip3Aa group performed with the MEGA 6.06 software. Branch lengths represent the number of substitutions per site of the multiple-sequence alignment as a measure of divergence. In addition, the bootstrapping values (number next to the nodes) indicates how many times out of 100% the same branch was observed when repeating the phylogenetic reconstruction on a re-sampled set of your data.