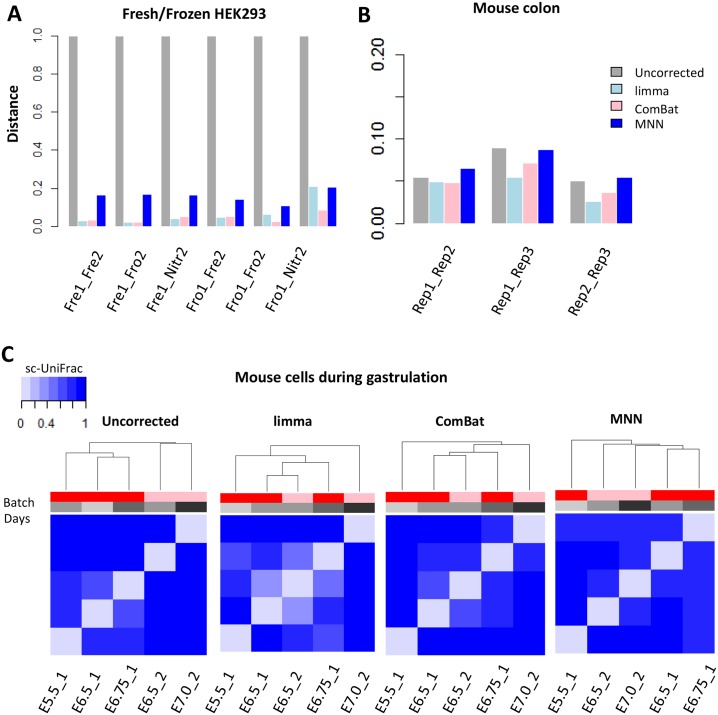
Fig 8. sc-UniFrac can benchmark batch effect removal approaches.
(A) sc-UniFrac distance calculated comparing uncorrected and batch-corrected scRNA-seq datasets of HEK293 cells fresh, frozen at −80 °C, or liquid nitrogen flash frozen performed in two different batches (GSE85534) [35]. ComBat, limma, and MNN were used for batch correction. (B) sc-UniFrac distance calculated similar to panel A for technical replicates of the mouse colonic epithelium scRNA-seq data (GSE102698). (C) Hierarchical clustering by sc-UniFrac of uncorrected or batch-corrected scRNA-seq data depicting murine gastrulation from two different studies [36,37]. A gradation of similarity, and hence clustering, was expected over developmental times from the earliest development stage (E5.5) to the latest stage (E7.5). Data from GSE100597; http://gastrulation.stemcells.cam.ac.uk/scialdone2016. E, embryonic day; Fre, fresh; Fro, frozen at −80 °C; HEK293, human embryonic kidney 293; Nitr, liquid nitrogen flash frozen; scRNA-seq, single-cell RNA sequencing.