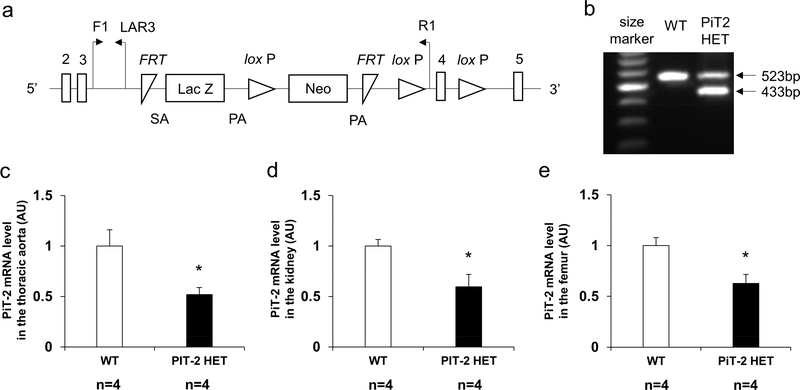
Figure 1 |. Genomic design of a knock-out first allele and characterization of PiT-2 HET mice.
(a) Presentation of the knockout-first allele. (b) Expression of genomic DNA determined by PCR in WT and PiT-2 HET mice. PiT-2 mRNA level in the (c) aorta, (d) kidney, (e) and femur. Three samples from each genotype were used for quantitative RT-PCR and six samples for biochemical determination. Data are expressed as mean ± SEM and compared by unpaired t-test. A two-tailed P<0.05 was considered statistically significant. *P<0.05 versus WT. AU, arbitrary unit; bp, base pair; F1, forward primer recognition site; FRT, flippase recognition target; LAR3, ligation amplification reaction. Neo, neomycin phosphotransferase; PA, polyadenylation; PiT-2 HET, PiT-2 heterozygous knockout; R1, reverse primer recognition site; SA, splice acceptor; WT, wild-type.