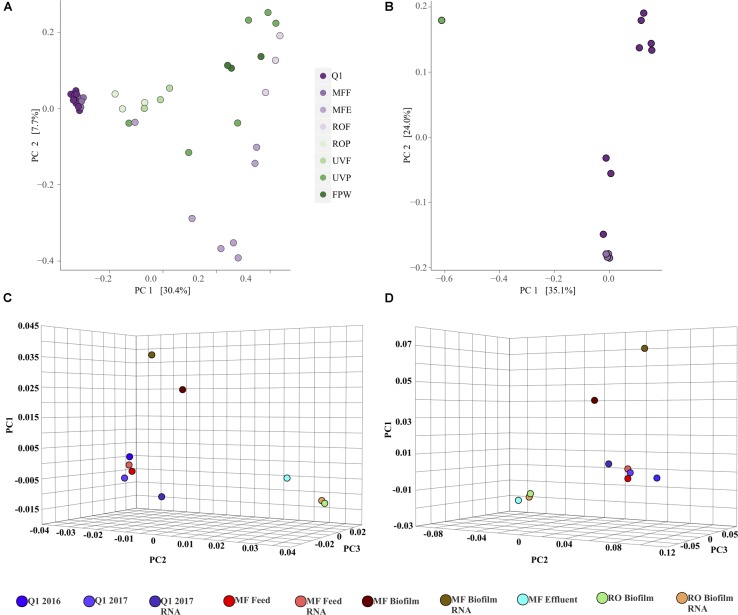
FIGURE 4.
Principal coordinate ordination, produced using an unweighted UniFrac distance matrix for the bacteria/archaea (A) or eukaryotes (B) from rRNA gene sequence data. Axis values represent percent variance explained by the ordination. Principal component ordination of the bacteria (C) or antibiotic resistance genes (D) produced using a Bray-Curtis distance matrix within the CosmosID web application. Additional information on the specific types of ARGs used in the ordination of 5d can be found in Supplementary Table S2.