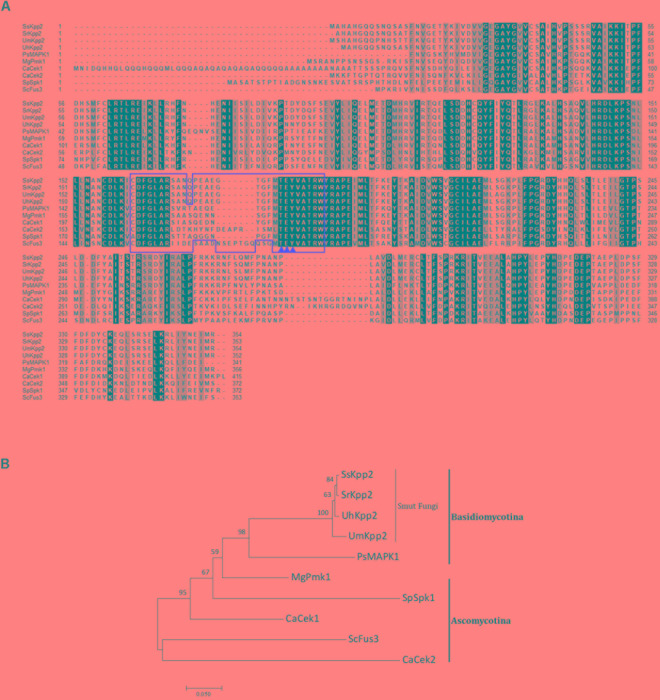
FIGURE 1.
Amino acid sequences arrangement and phylogenetic analysis with SsKpp2 protein and its orthologs. (A) Amino acid sequences arrangement and phylogenetic analysis with SsKpp2 protein and its fungal orthologs: S. reilianum SrKpp2 (CBQ73711), U. maydis UmKpp2 (AAF15528), U. hordei UhKpp2 (CCF52019), Puccinia striiformis PsMAPK1 (ADL57241), M. oryzae MgPmk1 (AAC49521), C. albicans CaCek1 (XP_715542) and CaCek2 (AAG43110), S. pombe SpSpk1 (NP_594009), and S. cerevisiae ScFus3 (NP_009537). The black and gray shadow denote identical and conserved residues, respectively. The red boxes and three red triangles represent STKc_MAPK domains and predicted dual phosphorylation lip sequences, respectively. (B) Phylogenetic analysis of Kpp2 othorlogs as listed in (A). The tree is calculated with Neighbor-Joining method (Saitou and Nei, 1987) using MEGA 7 (Kumar et al., 2016). Numbers beside each node indicate a percentage of 1000 bootstrap replications, computed using the Poisson correction method (Zuckerkandl and Pauling, 1965).