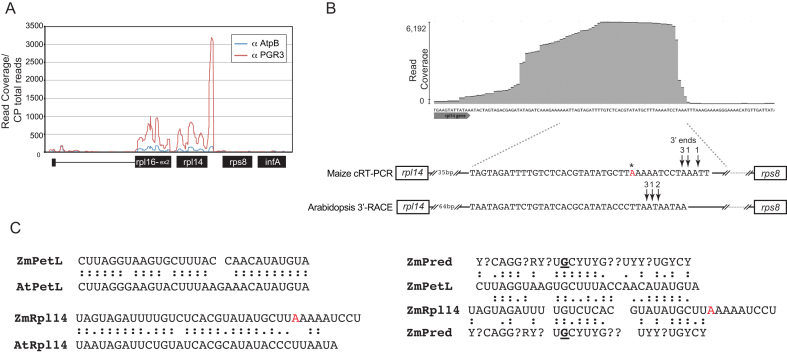
Figure 4.
RIP-seq analysis showing that Zm-PGR3 associates with RNA from the rpl14-rps8 intergenic region in vivo. Antibody to Zm-PGR3 was used for immunoprecipitation from maize chloroplast stroma. RNA purified from the immunoprecipitate was analyzed by deep sequencing. An immunoprecipitation with antibody to AtpB (a subunit of the chloroplast ATP synthase) served as a negative control. (A) Sequence read counts in the rpl14 region. Read counts were normalized to the total number of reads mapping to the chloroplast genome. A genome-wide view of the data is shown in Supplementary Figure S3. (B) Position of the PGR3-binding site and PGR3-dependent 3′-ends in the rpl14-rps8 intergenic region. A screen capture of an excerpt of the PGR3 RIP-seq data displayed with the Geneious browser is shown at top. The sequence is expanded below, where the positions of the PGR3-dependent 3′-ends as mapped by circular RT-PCR (maize) or 3′-RACE (Arabidopsis) are marked. The number of clones representing each 3′-end is indicated. The asterisk marks a difference in the sequences we obtained from that in the reference maize chloroplast sequence (GENBANK Accession NC_001666). (C) Comparison of PGR3’s binding sites near petL and rpl14. Alignments between the orthologous sites in maize (Zm) and Arabidopsis (At) are shown at left. An alignment between the maize petL and rpl14 sites and the Zm-PGR3 binding site as predicted by the PPR code (ZmPred) is shown to the right. The code prediction was based on the nucleotide specificities according to the published PPR code (8). PPR motifs that lacked canonical amino acids at the specificity-determining positions are marked with question marks. The binding site prediction for Arabidopsis PGR3 is similar, with the exception of the position in bold underline near the center of the protein, which is predicted to be a pyrimidine in Arabidopsis.