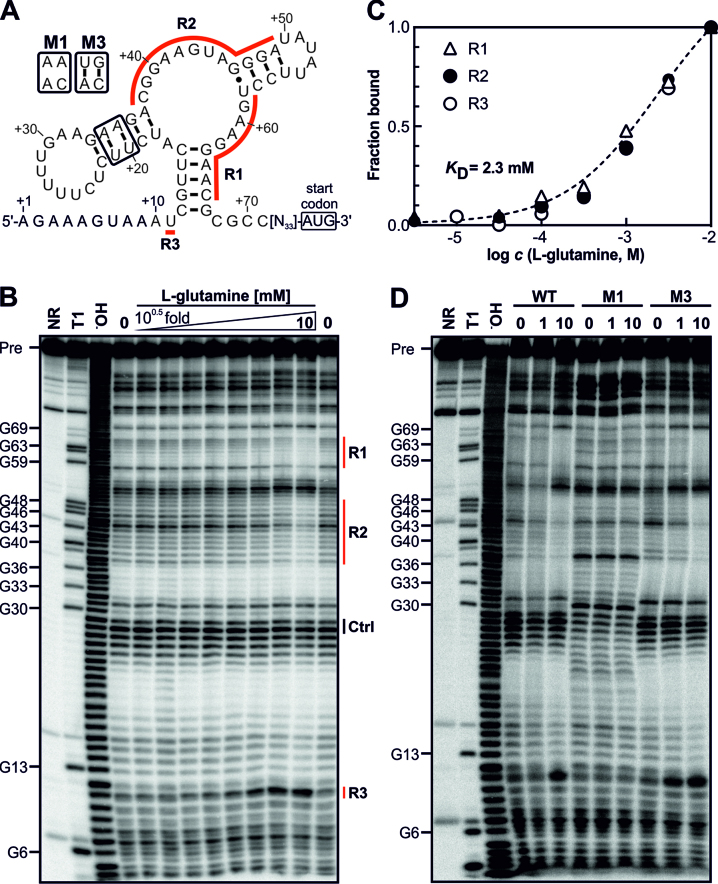
Figure 2.
The gifB 5′ UTR of Synechocystis harbors a glutamine-binding aptamer. (A) Predicted secondary structure of the glnA aptamer upstream of the gifB gene. The nucleotide positions refer to the transcriptional start site of the gifB gene (+1, ref. (21)). R1–3 label the regions for which structural modulation was observed (see panel B). The secondary structure is based on the secondary structure model of glnA motif RNAs published previously (42). (B) In-line probing analysis using a 5′ 32P-labeled gifB 5′ UTR. Precursor RNA (Pre) was incubated for 2 days at 23°C without a ligand (0) or in presence of increasing concentrations of L-glutamine (3.2 μM to 10 mM, half-log dilutions). The same RNA was also partially treated with Na2CO3, which mediates alkaline degradation (−OH) or with RNase T1, which cleaves after G residues (T1). A non-treated RNA served as control (NR, no reaction). (C) Densitometric evaluation of the bands corresponding to regions R1–3 whose intensities changed due to the presence of glutamine (marked by red lines in panel B). The intensities were normalized to bands which did not show glutamine-dependent intensity changes (Ctrl). (D) In-line probing using mutated RNAs. The NR, T1 and −OH samples are shown for the WT RNA.