Figure 2.
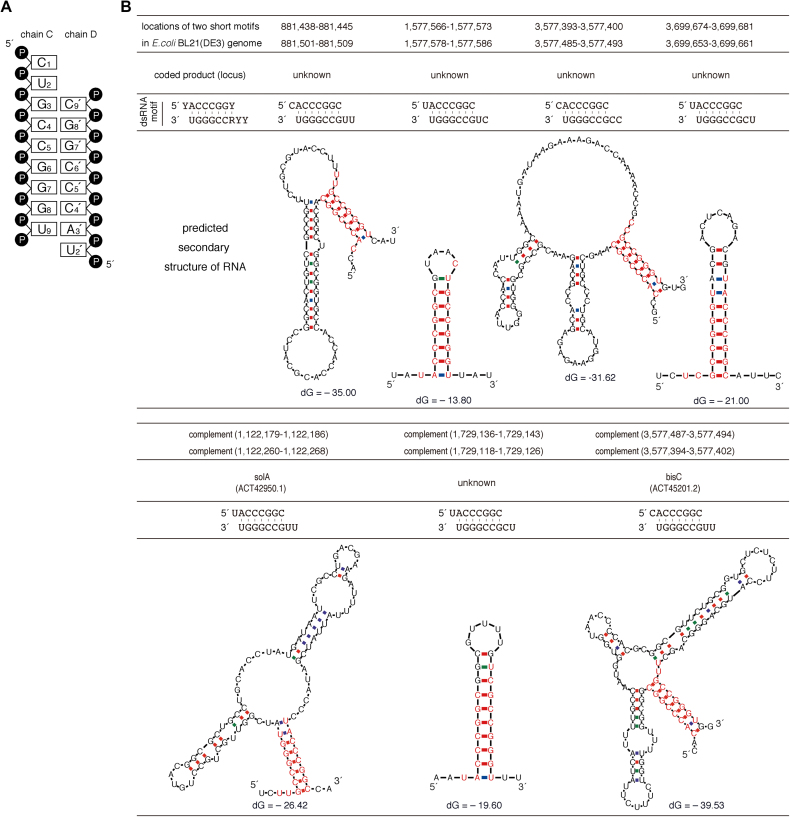
Identification of Escherichia coli-derived nucleotide sequences analogous to the dsRNA detected in the cpzA3H crystal structure. (A) Diagram of the dsRNA secondary structure modeled in the crystal structure. (B) Seven of our identified nucleotide sequences and their putative RNA secondary structures are shown. The dsRNA sequences identified in the crystal structure are shown as the ‘dsRNA motif’, consisting of 5′-YYRCCGGGU and 5′-YACCCGGY (Y, pyrimidine—C or U; R, purine—G or A). Using an ‘in-house’ program, the locations of these two nucleotide sequences were determined in the complete genome of E. coli BL21(DE3), which was used for cpzA3H protein expression. Based on the genome sequences around the identified positions, the RNA secondary structures were predicted with the mfold program. The nucleotides analogous to the dsRNA motif are highlighted in red. No dsRNA motifs were identified in the GST-cpzA3H expression plasmid.