Figure 1.
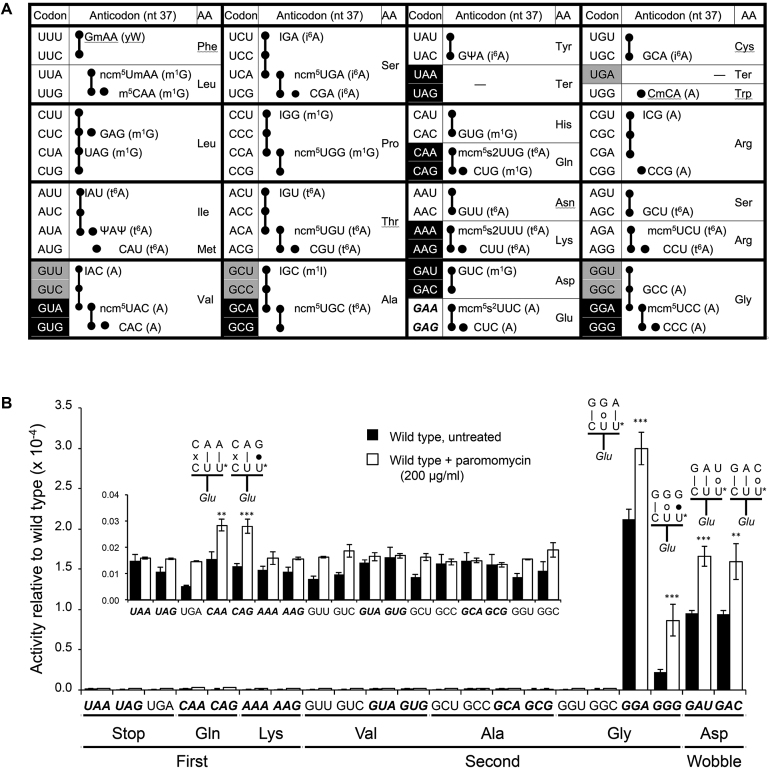
Error frequencies in yeast can vary by 100-fold as measured by 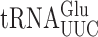 on various near-cognate codons. (A) Genetic code and identity of cytoplasmic tRNAs in Saccharomyces cerevisiae. The codons recognized by each tRNA are indicated by black circles connected by bars; the tRNAs are identified by anticodon and position 37 nt and the encoded amino acid. The Glu codons decoded by
on various near-cognate codons. (A) Genetic code and identity of cytoplasmic tRNAs in Saccharomyces cerevisiae. The codons recognized by each tRNA are indicated by black circles connected by bars; the tRNAs are identified by anticodon and position 37 nt and the encoded amino acid. The Glu codons decoded by 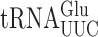 are in italics; near-cognate codons for
are in italics; near-cognate codons for 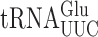 are highlighted in black and synonymous non-cognates in gray. (B) The activity of E537 mutants of β-galactosidase expressed with or without treatment with the antibiotic paromomycin. Statistical significance of the effect of paromomycin is shown (*, P-value < 0.05; **, P-value < 0.01; ***, P-value < 0.001). For mutants showing a significant change the codon–anticodon complexes predicted for corresponding misreading events are shown (the upper line represents the codon, the lower the anticodon). Vertical lines represent Watson–Crick pairs, filled circles canonical wobble pairs and open circles non-Watson–Crick pairs that have been shown to mimic Watson–Crick geometry.
are highlighted in black and synonymous non-cognates in gray. (B) The activity of E537 mutants of β-galactosidase expressed with or without treatment with the antibiotic paromomycin. Statistical significance of the effect of paromomycin is shown (*, P-value < 0.05; **, P-value < 0.01; ***, P-value < 0.001). For mutants showing a significant change the codon–anticodon complexes predicted for corresponding misreading events are shown (the upper line represents the codon, the lower the anticodon). Vertical lines represent Watson–Crick pairs, filled circles canonical wobble pairs and open circles non-Watson–Crick pairs that have been shown to mimic Watson–Crick geometry.