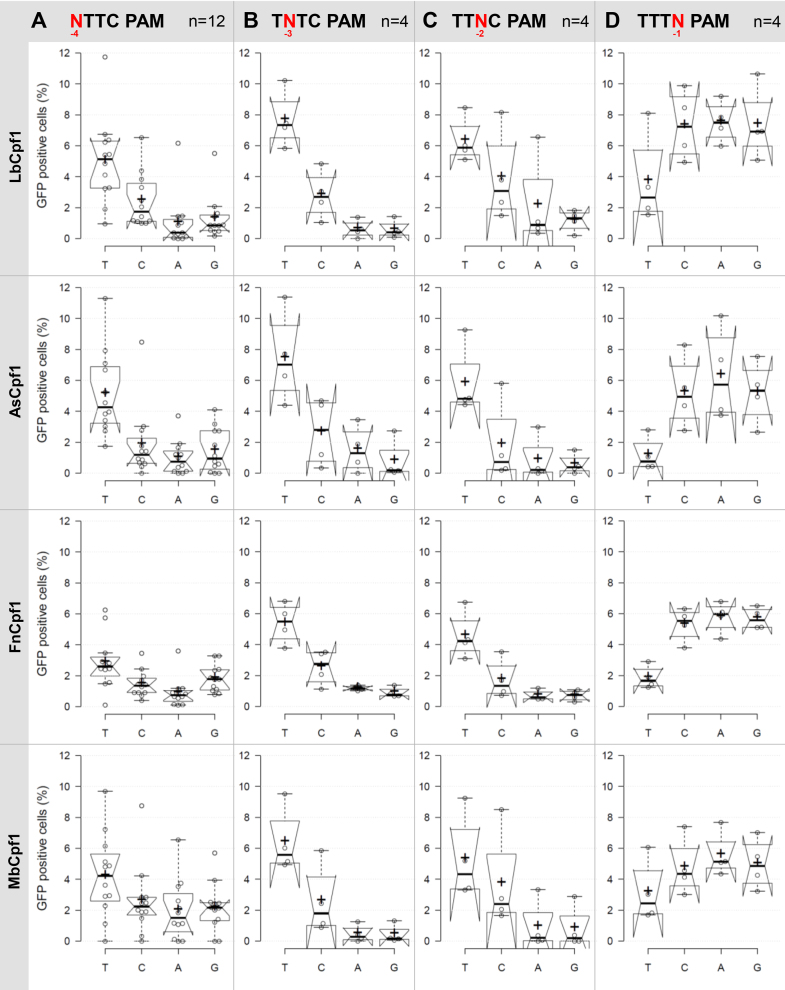
Figure 5.
PAM requirement of Lb-, As-, Mb- and FnCpf1 nucleases. Percentages of GFP positive cells counted above the background level, resulting from the action of various nucleases on targets with different PAM sequences. Four or twelve (indicated as n) randomly picked targets with different PAM sequences (A) NTTC PAM sequence, (B) TNTC PAM sequence, (C) TTNC PAM sequence, (D) TTTN PAM sequence) cloned into the pGF-ori-FP vector were tested using the GFxFP assay (19). The target vectors along with the corresponding nuclease vector were transfected into N2a cells and GFP positive cells were counted 2 days after transfection. All samples were also cotransfected with an mCherry expression vector to monitor the transfection efficiency and the GFP signal was analyzed within the mCherry positive population. The background fluorescence was assessed by using a crRNA-less, inactive AsCpf1 nuclease expression vector as negative control and was subtracted from each sample. Three parallel transfections were made for each case. Tukey-type notched boxplots by BoxPlotR (53): center lines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; notches indicate the 95% confidence intervals for medians; crosses represent sample means; data points are plotted as open circles that correspond to the different targets tested. See also Supplementary Figures S9–S12.