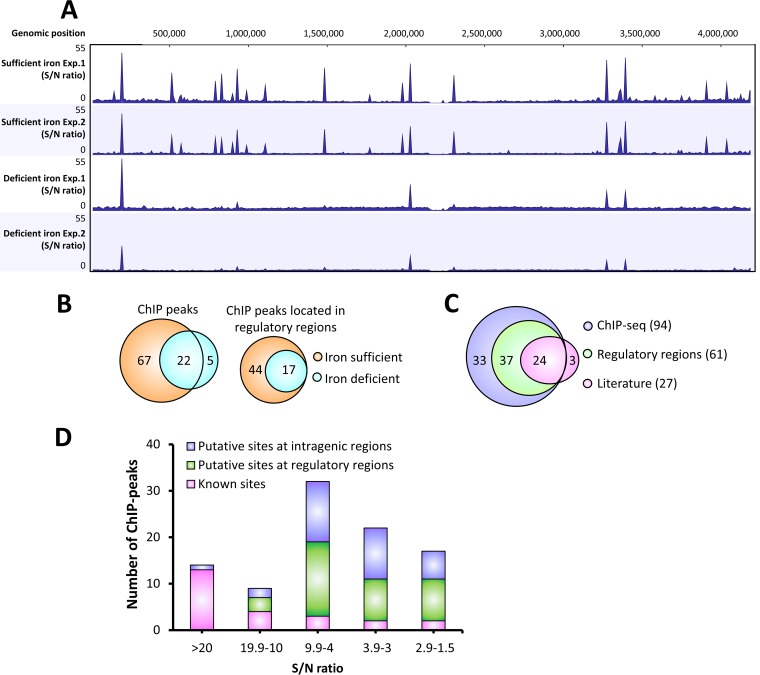
FIG 1.
Overview of Fur-binding profiles across the B. subtilis genome under varied iron conditions. (A) ChIP-seq data of B. subtilis Fur-dependent binding under iron-sufficient and iron-deficient conditions. Most of the ChIP peaks showed higher occupancy under iron-sufficient conditions. Two biological replicates were included for each growth condition (Exp. 1 and Exp. 2). S/N denotes the signal-to-noise ratio for peak calling. (B) Most of the ChIP peaks (22 out of 27) identified under iron-deficient conditions overlap those detected under iron-sufficient conditions. The majority of peaks (61 out of 94) are located in regulatory regions. (C) Most of the known Fur binding sites from the literature (24 out of 27) are identified by ChIP-seq, with three exceptions (see the text). Among all the new identified Fur binding sites, 33 sites are located in intragenic regions, while 37 sites are located in regulatory regions. (D) Distribution of ChIP peaks relevant to the S/N ratio. The highest S/N ratio of each ChIP peak among all four samples was used for this analysis.