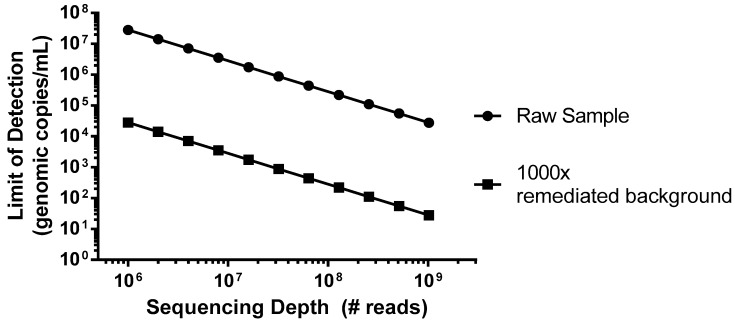
Figure 1.
Predicted sensitivity for porcine circovirus (PCV) at a given sequencing depth, with and without incorporation of steps to reduce cellular nucleic acid background. Calculated deep sequencing results of a sample composed of 1 × 107 CHO cells/mL (2.4 × 109 bp genome) spiked with PCV type 2 (PCV-2) (1.7 × 103 bp genome). For a given sequencing depth, the viral concentration at which a single viral read is expected to be obtained is shown. For example, if we would like to ensure sensitivity for PCV-2 above 1 × 106 copies/mL, we would need to generate at least 1 × 107 total reads. The effect of reducing background CHO cell DNA by 1000-fold is shown in the graph by closed squares. Under these conditions, we can expect a similar sensitivity with only 3 × 105 total reads.