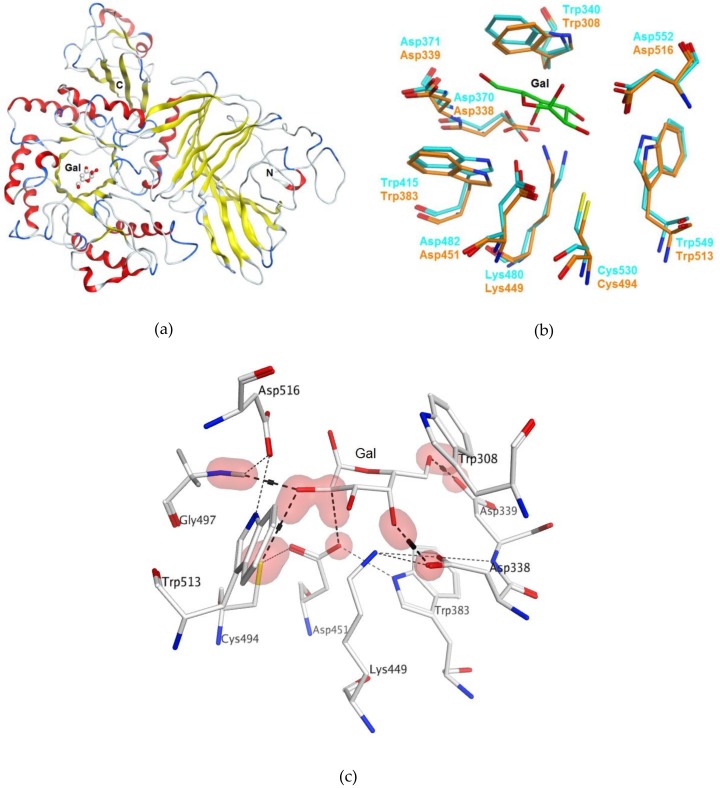
Figure 1.
Homology model of α-PsGal three-dimensional (3D) structure generated using X-ray structure of the α-galactosidase of Lactobacillus acidophilus (PDB ID 2XN2) as a template: (a) 3D-model of α-PsGal structure in a ribbon diagram representation: α-helixes (red), β-strands (yellow), coils (white), and turns (blue); (b) superimposition of the α-PsGal homology model (orange) with template active sites (turquoise); D-galactose is shown by sticks (green); and (c) the binding site of D-galactose in the active center of α-PsGal homology model. Hydrogen-bond contacts were determined using the Protein Contacts module of Molecular Operating Environment version 2018.01 (MOE) program (Chemical Computing Group ULC: 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2018) and are shown with a dashed line.