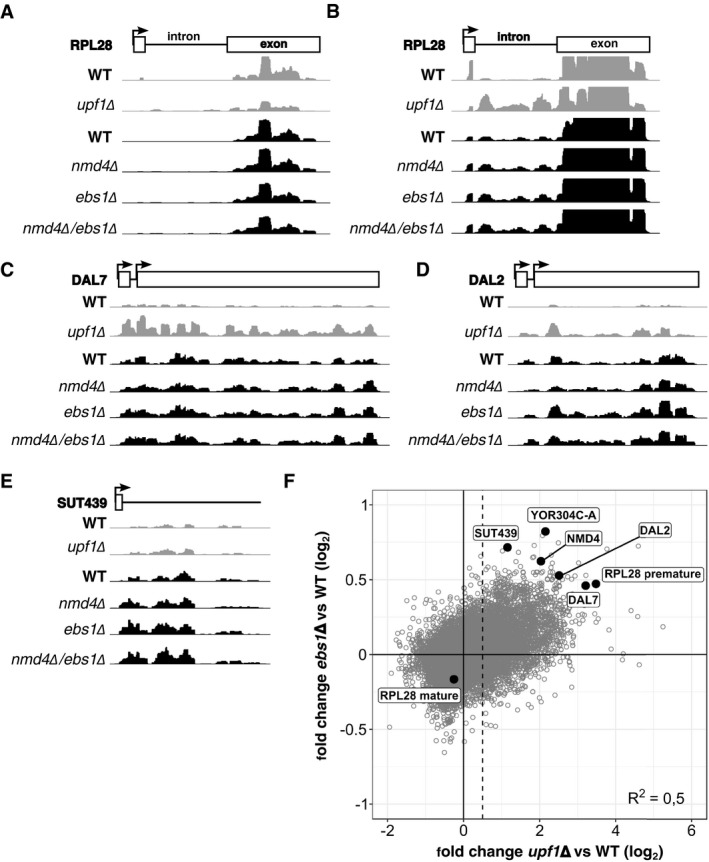
Figure EV3. Deletion of NMD4 and EBS1 stabilize a set of transcripts that is also stabilized in the absence of UPF1 .
-
A–EExamples of NMD substrates sequencing profiles in WT, upf1∆, nmd4∆, ebs1∆ and nmd4∆/ebs1∆ experiments. NMD substrates belong to different classes, intron containing (RPL28; A and B, different scaling to show intron signal), uORF (DAL7, DAL2; C and D) and non‐coding RNA (SUT439; E). Profiles were normalized using the samples median counts.
-
FScatter plot of transcript log2 fold change in upf1∆ against ebs1∆. The dashed line represents the limit over which RNA was considered as stabilized, with a log2 value of 0.5 (1.4 fold change).
