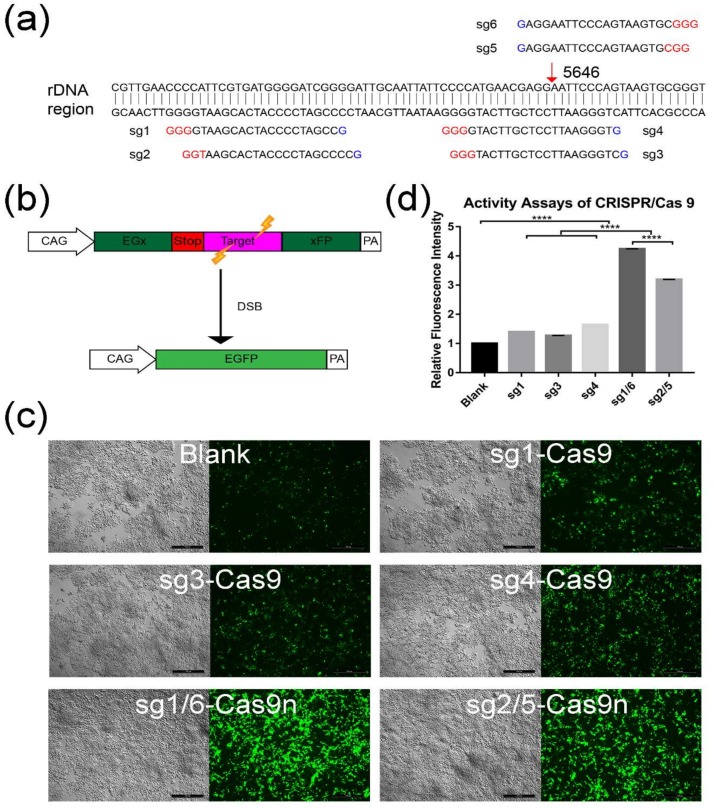
Figure 1.
Activity assay for different sgRNA-CRISPR/Cas9 groups. (a) Binding position of six different sgRNAs. Red arrow indicates the targeting site in the rDNA region. (b) Schematic illustration of activity assays by using the pCAG-EGx-Target-xFP system. The EGFP (enhanced green fluorescence protein) cDNA was divided to two parts, EGx and xFP, in which the “x” represents a repeat fragment of 482 bp. The two parts were separated by the target region (a) 77 bp fragment cloned from the rDNA region) along with the stop code. When the sgRNA-CRISPR/Cas9 worked and introduced double strand breaks (DSB), the EGFP cassette was reconstituted by either HDR or SSA. Thus, the green fluorescence could be observed. (c) The activity assay for sgRNA-CRISPR/Cas9 was conducted by co-transfecting HEK 293T cells with pCAG-EGx-Target-xFP and sgRNA-CRISPR/Cas9 or sgRNA-CRISPR/Cas9n plasmids (named sg1-Cas9n, sg2-Cas9n, sg5-Cas9n, sg6-Cas9n sg1-Cas9, sg3-Cas9, and sg4-Cas9). sg1/6-Cas9n group means co-transfection with sg1-Cas9n and sg6-Cas9n. In the same way, sg2/5-Cas9n group means co-transfection with sg1-Cas9n and sg6-Cas9n. Blank represented cells only transfected with the plasmid pCAG-EGx-Target-xFP. After 72 h, the green fluorescence of transfected cells was visualized. Scale bar: 500 µm. (d) GFP fluorescence intensity of each group was quantified and analyzed with Image J. GraphPad Prism 7.0 was used for calculation and statistics analysis. The relative fluorescence intensity was normalized to blank. (****, p < 0.0001).